Abstract
Background
Accurate and rapid diagnosis of Clostridium difficile infection (CDI) is critical for effective patient management and implementation of infection control measures to prevent transmission.
Objectives
We updated our previous meta-analysis to provide a more reliable evidence base for the clinical diagnosis of Xpert C. difficile (Xpert C. difficile) assay.
Methods
We searched PubMed, EMBASE, Cochrane Library, Chinese National Knowledge Infrastructure (CNKI), and the Chinese Biomedical Literature Database (CBM) databases to identify studies according to predetermined criteria. STATA 13.0 software was used to analyze the tests for sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and area under the summary receiver operating characteristic curves (AUC). QUADAS-2 was used to assess the quality of included studies with RevMan 5.2. Heterogeneity in accuracy measures was tested with Spearman correlation coefficient and chi-square. Meta-regressions and subgroup analyses were performed to figure out the potential sources of heterogeneity. Model diagnostics were used to evaluate the veracity of the data.
Results
A total of 26 studies were included in the meta-analysis. The pooled sensitivity (95% confidence intervals [CI]) for diagnosis was 0.97(0.95–0.98), and specificity was 0.96(0.95–0.97). The AUC was 0.99 (0.98–1.00). Model diagnostics confirmed the robustness of our meta-analysis’s results. Significant heterogeneity was still observed when we pooled most of the accuracy measures of selected studies. Meta-regression and subgroup analyses showed that the sample size and type, ethnicity, and disease prevalence might be the conspicuous sources of heterogeneity.
Conclusions
The up-to-date meta-analysis showed the Xpert CD assay had good accuracy for detecting CDI. However, the diagnosis of CDI must combine clinical presentation with diagnostic testing to better answer the question of whether the patient actually has CDI in the future, and inclusion of preanalytical parameters and clinical outcomes in study design would provide a more objective evidence base.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Clostridioides (Clostridium) difficile infection (CDI) is the leading cause of healthcare-associated infections in the USA and is responsible for approximately 15,000 deaths annually in the USA [1, 2]. It accounts for 15 to 25% of healthcare-associated diarrhea cases in all healthcare settings [3]. Acquisition of C. difficile as a healthcare-associated infection (HAI) is associated with increased morbidity and mortality. It has been estimated that the length of CDI-associated hospital stays has increased and the average cost per case for HAI is more than $30,000 which is 1.5 times the cost of community-associated CDI (CAI) [4].
The clinical signs and symptoms presented by CDI are highly nonspecific, making it difficult to differentiate CDI from non-CDI, including non-CDI diarrhea in a C. difficile-colonized patient [5]. Therefore, accurate diagnosis of CDI is critical for effective patient management and implementation of infection control measures to prevent transmission [6]. Diagnostic tests for C. difficile are classified as tests for C. difficile products (GDH, TcdA, and TcdB), toxigenic culture methods (TC), cell cytotoxicity neutralization assay (CCNA), toxin detection by enzyme immunoassays (EIA), and detection of toxin genes by nucleic acid amplification tests (NAATs). However, the best practices for laboratory diagnosis of CDI remain controversial [7]. The anaerobic toxigenic culture (TC) and culture cytotoxicity neutralization assay (CCNA) were often used as the laboratory reference tests for detecting C. difficile. Unfortunately, both tests are slow and labor-intensive [8]. EIAs for toxins A and B are rapid and relatively inexpensive, but it was ultimately demonstrated that EIAs cannot be used as stand-alone tests due to their low sensitivity [9]. Although the accurate and rapid diagnosis of CDI is essential for effective and timely treatment, this remains an unmet clinical need.
Currently, several NAATs have been cleared by the Food and Drug Administration (FDA) [10] and supported by recent guidelines by the American Society of Microbiology [11]. We previously published a meta-analysis to evaluate the Xpert C. difficile system (Cepheid, USA), a platform that detects the toxin B gene (tcdB), the CDT component A gene (cdtA), and a deletion within the LCT regulatory gene tcdC to putatively identify “hypervirulent” RT 027 [12]. These strains have been shown to produce a large amount of toxins in vitro and are associated with erythromycin and newer fluoroquinolones resistance. The Xpert C. difficile assay is among the simplest to perform and, with a turnaround time of about 1 h, is also the most rapid of the NAATs available. Since the publication of the previous meta-analysis, four new researches including 1141 patients that evaluated the diagnostic accuracy of Xpert C. difficile assay have been published.
As the latest 2018 guidelines from the IDSA recommend that NAATs can be used as a stand-alone diagnostic test in cases where there are pre-agreed institutional criteria for patient stool submission [13], we identified the need to update our 2017 meta-analysis to provide a more reliable evidence base for the clinical application of Xpert C. difficile assay, and the results are presented in this report.
Materials and methods
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines in our study.
Literature search
PubMed, EMBASE, Cochrane Library databases, Chinese National Knowledge Infrastructure (CNKI), and Chinese Biomedical Literature Database (CBM) were searched from July 2016 to the end of April 2020 without language restrictions by two investigators (YY Bai and YL Wang). The search terms used were as follows: Clostridium difficile AND (Xpert C. difficile OR molecular diagnostic techniques). Reference lists from included studies were also searched. We reviewed and included our previous search and added all relevant articles, focusing on the time after the previous search.
Study criteria
We searched the literature using the following predetermined inclusion criteria. Studies evaluating Xpert CD as a diagnostic test for CDI were eligible for inclusion if the studies (1) described original research; (2) performed stool samples analyses from human patients, either children or adults; (3) compared Xpert CD to a reference method — either CCNA or anaerobic TC; and (4) had extractable data to fill the 4 cells of a 2 × 2 table for diagnostic tests (true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN)).
Relevant publications were excluded if they were duplicated articles, letters without original data, animal studies, case reports, editorials, and reviews. Studies with fewer than 20 samples were also excluded to reduce selection bias. Articles that contain data from infants were excluded because infants rarely develop clinical infection.
Data extraction
Two investigators (YY Bai and YL Wang) extracted data from full text of the included studies independently. Disagreements were resolved by consensus. Information was extracted on the first author, publication year, country where the study was conducted, sample size, reference tests the diagnosis used, the number of TP, the number of FP, the number of FN, and the number of TN. These were summarized as sensitivity, TP/(TP + FN); specificity, TN/(TN + FP); and prevalence, (TP + FN)/(TP + FN + TN + FN).
Quality of study reports
We applied the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) to assess the quality of included studies (http://www.bris.ac.uk/quadas/), an updated version of the original software [14].
Statistical analysis
Accuracy estimates
Meta-analyses were performed using two software programs: STATA 13.0 (Stata Corporation, Texas, USA) and Cochrane RevMan 5.2. Sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), forest plots, and summary receiver operating characteristic (SROC) curves were analyzed with the “MiDAS” module for STATA 13.0, based on the random model effect. Quality of studies was assessed with RevMan 5.2.
Heterogeneity
We used chi-square test and I2 (p < 0.05 and I2 > 50% indicated significant heterogeneity) to identify heterogeneity. Heterogeneity was evaluated using the methods detailed in our previous study [15]. To assess the potential sources of heterogeneity, we further performed subgroup analyses and meta-regressions according to the descriptions of the included studies. Model diagnostics were used to evaluate the veracity of the data. Extreme outliers and highly influential cases were reevaluated and corrected as described above if appropriate. Deeks’ funnel plot asymmetry test was performed to investigate the publication bias, with p < 0.10 showing significant publication bias, which is an important concern for meta-analyses of diagnostic accuracy [16].
Results
Characteristics of the selected studies
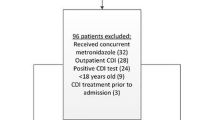
A flow chart of the study selection process is shown in Fig. 1. A total of 280 potentially relevant citations were identified from all searches. Finally, according to the inclusion and exclusion criteria, since the publication of our previous meta-analysis, four new Asian-based studies involving 1141 additional patients were included in this updated meta-analysis. Because diagnostic tests performed with different reference methods occurred in the same article, 26 independent studies (including 10,493 samples) were defined in the meta-analysis. Table 1 shows the characteristics of these included studies [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40]. Most of the studies were prospective in design, but only 10 studies were blind (data not shown).
Quality assessment
A quality assessment of all the included articles is illustrated in Fig. 2. In conclusion, patient selection provided the most high-risk bias and high-risk applicability concerns. Nearly half of the included articles were at either high-risk or unclear-risk bias in “patient selection” and “flow and timing” domains of QUADAS-2 due to the lack of detail regarding timing, inconsecutive, or nonrandom patient selection and blinding. A total of 13 (54%) studies were at low risk, 7 studies (29%) were of unclear risk, and 4 studies (17%) were at high risk for patient selection bias. Most of the articles provided either low or unclear risk in the index test and reference standard bias domains. Regarding applicability, half of the articles were at high risk for patient selection; however, most of the articles (n = 22, 91.7%) were in the low risk of index test and reference standard domains.
Diagnostic accuracy
Results are given as values (95% CI). Using a random-effects model, the results were as follows: sensitivity 0.97(0.95–0.98), I2 = 74.9%; specificity 0.96(0.95–0.97), I2 = 87.4% (Fig. 3); PLR 23.81(17.93–31.61), I2 = 82.27%; NLR 0.03 (0.02–0.05), I2 = 71.40%; DOR 784.85 (440.25–1399.16), I2 = 100%; and AUC 0.99 (0.98–1.00) (Fig. 3).
Forest plots of the pooled sensitivity and specificity and SROC curve of Xpert CD for detection of CDI. a Forest plots of the pooled sensitivity and specificity. Each solid square represents an individual study. Error bars represent 95% CI. Diamond indicates the pooled sensitivity and specificity for all of the studies. b SROC curve
To create an overall index of effect, the likelihood ratio scatter matrix was utilized. The paired likelihood ratios are within the areas that are typically used to indicate high clinical validity (+ LR of > 10 and -LR of < 0.1), the expert panel described this as a “substantial” effect, and the error bands of the estimate (as represented by the crosshairs on the summary diamond) do not cross into other quadrants (Fig. 4).
Robustness tests
Goodness of fit and bivariate normality analyses (Fig. 5a, b) showed the bivariate model was moderately robust. Influence analysis and outlier detection identified only two outliers [18, 24] (Fig. 5c, d). Instructively, conducting the same analyses after excluding the two outliers did not significantly change the overall results (Table 2). Finally, the Deeks’ funnel plot asymmetry test was conducted to assess publication bias in this study (Fig. 6), which suggested statistically significant publication bias (p = 0.04).
Heterogeneity
There was substantial heterogeneity for all the statistical measures. The heterogeneity test results of sensitivity and specificity are illustrated in the forest plots (Fig. 3). The Spearman correlation coefficient between the logit of sensitivity and logit of 1-specificity was used to assess the threshold/cutoff effect. The Spearman correlation coefficient (p value) in diagnosis of CDI was 0.237 (p = 0.244). This indicated that the heterogeneity might not be due to threshold/cutoff effect. To assess for causes of variations other than threshold, we further performed meta-regressions and subgroup analyses according to the descriptions of the included studies.
Meta-regression and subgroup analyses
Meta-regression analyses were performed to further investigate the potential sources of inter-study heterogeneity (Fig. 7). Notably, the results showed that the sample size, sample type, the type of ethnicity, prevalence, and whether blinded all were conspicuous sources of heterogeneity, and most of these variables had a more significant effect on specificity than sensitivity. In addition, subgroup analyses based on these variables were performed; the related parameters of which, including pooled sensitivity, specificity, PLR, NLR, and DOR for each subgroup, are also listed in Table 2.
Discussion
The incidence and severity of CDI have been increasing significantly worldwide with associated morbidity, mortality, and healthcare costs [41]. A rapid and accurate diagnosis is essential to guide the treatment and to prevent transmission. It has been shown that rapid diagnosis positively impacts on patient’s care by reducing delays in initiation of isolation and treatment for confirmed CDI cases [42]. The Xpert CD assay is now implemented in many countries due to its shorter turnaround time, thus a more effective procedure. The most significant advantage of the Xpert CD assay is its rapidity and simplicity. According to the Society for Healthcare Epidemiology of America and the Infectious Diseases Society of America guidelines, “...PCR testing appears to be rapid, sensitive, and specific and may ultimately address testing concerns. More data on utility are necessary before this methodology can be recommended for routine testing.”
To provide much more evidence-based results for the utility of this assay in routine testing, we performed a meta-analysis to comprehensively evaluate the overall diagnostic accuracy of the Xpert CD assay in detecting CDI compared with reference tests in 2017. Our previous meta-analysis concluded that the Xpert CD assay had good accuracy for detecting CDI (sensitivity 0.97(0.95–0.99); specificity 0.95(0.94–0.96); PLR 21.41(16.66–27.52); NLR 0.03 (0.02–0.05); DOR 762.13(401.82–1445.52); and AUC 0.99(0.97–0.99)). Nonetheless, we found significant heterogeneity for diagnostic parameters among the studies analyzed and the considerable heterogeneity among the results remained unexplained even after subgroup analysis.
Since the publication of our previous report, several newer studies have been published to evaluate the diagnostic accuracy of Xpert CD assay, and four new researches including 1141 cases meet the inclusion criteria for the meta-analysis. We implemented this meta-analysis to offer an up-to-date and comprehensive analysis. A total of 26 independent studies (including 10,493 samples) were finally included in this updated meta-analysis. Moreover, further advantageous features introduced into the analysis methodology included: (1) influence analysis and outlier detection were performed to evaluate the robustness of the tests; (2) meta-regression and subgroup analyses were conducted to evaluate the prospective sources of heterogeneity; and (3) the Deeks’ funnel plot asymmetry test was conducted to assess publication bias.
Since meta-analytic procedures model the summary, estimations are only as trustworthy as the implemented models. Thus, evaluations of model diagnostics (e.g., goodness of fit, influence analysis, outlier detection) are important [43], while there were only two influential outliers through influence analysis and outlier detection. After we excluded the two outliers and then made the same analyses for the leaving studies, we found that the overall results did not change significantly (Table 2), confirming the robustness of our meta-analysis. However, the Deeks’ funnel plot asymmetry test showed that there was statistically significant publication bias (p = 0.04).
As with our previous meta-analysis, this up-to-date meta-analysis still showed significant heterogeneity for diagnostic parameters among the studies analyzed. The Spearman correlation coefficient between the logit of sensitivity and logit of 1-specificity was not significant, indicating that the heterogeneity was not caused by threshold/cutoff effect. Since there were variations in sample characteristics and preanalytical and analytical procedures, heterogeneity can be assumed in diagnostic accuracy meta-analyses. However, unlike meta-analyses of treatment studies, there are no generally accepted measures of heterogeneity for diagnostic accuracy in meta-analyses [44]. Meta-regression can be used to identify and screen the main factors of heterogeneity, analyze the sources of heterogeneity, and provide guidance for further data collection in the future. It also provides the basis for future subgroup analysis.
Meta-regression and subgroup analysis in this meta-analysis were conducted to explore potential sources of heterogeneity. The meta-regression results showed that the sample size (> median 248 or < 248), sample type (fresh stool or frozen stool), the type of ethnicity (Asian or Caucasian), and prevalence (> 15% or < 15%) were conspicuous sources of heterogeneity, and we found that most of these variables had a greater influence on specificity than sensitivity in the inter-study heterogeneity (refer Fig. 7). It is important to be aware not only of the sensitivity and specificity of an assay but also of the CDI prevalence in the tested population, as the predictive values and hence the clinical utility of the assays depend on them [45]. Here the demographic characteristics (e.g., race, region) of the subjects in different regions are different, and the research levels in different countries are also inconsistent, so ethnicity (Asian or Caucasian) may be a potential source of heterogeneity. Subgroup analyses based on these variables were performed to test for causes of variations other than threshold effect. There were no significant heterogeneity for PLR (I2 = 23.39%, p = 0.01) and DOR (I2 = 21.93%, p = 0.19) when CDI prevalence of studies greater than 15% were pooled and that for sensitivity (I2 = 37.05%, p = 0.09), NLR (I2 = 13.8%, p = 0.31) and DOR (I2 = 0.00%, p = 0.69) when sample size of studies less than median sample size were pooled. There was moderate heterogeneity for sensitivity (I2 = 60.16%, p = 0.00) and NLR (I2 = 55.56%, p = 0.00) when studies using fresh stools tests were pooled. The results suggested that the CDI prevalence, sample size, and sample type could partly explain the heterogeneity.
The goal of this meta-analysis was to determine whether the Xpert CD assay had good diagnostic accuracy for CDI. This updated analysis showed this assay had “very good” diagnostic accuracy (sensitivity 0.97(0.95–0.98); specificity 0.96(0.95–0.97); PLR 23.81(17.93–31.61); NLR 0.03 (0.02–0.05); DOR 784.85(440.25–1399.16); and AUC 0.99 (0.98–1.00)). The performance of NAATs is commonly assessed using diagnostic accuracy measures for the presence of the C. difficile organism (e.g., sensitivity, specificity, PPV, and NPV). However, these measures may not directly link to the clinical definition of CDI or clinical outcomes, and some measures (e.g., PPV and NPV) are dependent on CDI prevalence in the patient population being tested [46]. Obviously, laboratory testing alone without considering the patient’s total clinical features is not appropriate for CDI diagnosis. Therefore, one of the limitations of our study was refined to be based only on the intermediate outcome of diagnostic accuracy for detecting the presence of the toxin genes; due to that, most of the included studies have limited evidence linking laboratory diagnosis with clinical outcomes.
Further, like other NAATs, one of the more important questions concern the clinical utility of Xpert CD assay is that it specifically detects the tcdB gene encoding the toxin and not the toxin itself. Therefore, asymptomatic carriers can be misdiagnosed as disease state patients if inappropriate testing is performed. To avoid overdiagnosis and overtreatment of toxigenic CDI by using the Xpert CD assay, it must be strictly limited to diarrheal stool specimen in patients without laxatives. The IDSA guideline states that if patients without laxatives meet the clinical criteria for CDI, and the laboratory does not test formed stools, either a stand-alone NAAT or an algorithm-based approach is acceptable [13]. The ASM guideline also endorses a stand-alone role of NAATs in CDI diagnosis. Indeed, the key to minimizing CDI overdiagnosis is appropriate patient selection, regardless of the diagnostic method used [47]. Preanalytic data are often used by clinicians when deciding if a patient should be tested for the presence of toxigenic C. difficile, such as history of antibiotic use or prior hospitalization, more than three times of diarrhea within 24 h, patient age, and residence in long-term care facilities. Consistently, our quality assessment analysis showed that “patient selection” provided the most high-risk bias and high-risk applicability concerns. All the included studies reported that the laboratory tested unformed or liquid stool; however, other details such as history of antibiotic use or prior hospitalization, and other known factors, were not included in the vast majority of diagnostic accuracy comparison studies, which made us fail to analyze the influence of preanalytic indicators on the diagnostic accuracy of Xpert CD assay. Moreover, even though the assay showed “very good” diagnostic accuracy, the absence of preanalytical factors also limits whether this meta-analysis can fully answer the critical question “Does this patient have CDI?” Therefore, it is crucial to note that the preanalytic variables of the clinical presentation should be taken into account with the interpretation of the diagnostic test result in future studies.
In conclusion, NAATs offer the combination of speed, sensitivity, high negative predictive value, and cost-effectiveness when used appropriately [7]. Stand-alone NAAT testing continues to be widely used for CDI. Although the up-to-date meta-analysis showed the Xpert CD assay had good accuracy for detecting CDI, however, it is clear that preanalytical factors are crucial for NAAT specifically. Hence, the diagnosis of CDI must combine patient presentation with diagnostic testing in the future. Low Xpert CD assay cycle threshold could indicate cytotoxicity assay positive patients and those with increased risk of mortality and possibly recurrence [48]. Lower mean quantification cycle values of NAATs could be a predictor of toxin presence in CDI [49]. Therefore, future studies are also needed to focus on the prediction of the disease severity and clinical outcomes. The inclusion of preanalytical parameters and clinical outcomes in study design would provide a more objective evidence base.
References
Schroeder LF, Robilotti E, Peterson LR, Banaei N, Dowdy DW (2014) Economic evaluation of laboratory testing strategies for hospital associated Clostridium difficile infection. J Clin Microbiol 52:489–496
Miller BA, Chen LF, Sexton DJ, Anderson DJ (2011) Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 32:387–390
Lessa FC, Winston LG, McDonald LC, Emerging Infections Program C. difficile Surveillance Team (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med 372:2369–2370
Barrett ML, Owens PL (2018) Clostridium difficile hospitalizations, 2011–2015. US Agency for Healthcare Research and Quality
McGlone SM, Bailey RR, Zimmer SM, Popovich MJ, Tian Y, Ufberg P, Muder RR, Lee BY (2012) The economic burden of Clostridium difficile. Clin Microbiol Infect 18:282–289
Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH (2018) Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48
Fang FC, Polage CR, Wilcox MH (2017) Point-counterpoint: what is the optimal approach for detection of Clostridium difficile infection? J Clin Microbiol 55:670–680
Crobach MJT, Baktash A, Duszenko N, Kuijper EJ (2018) Diagnostic guidance for C. difficile infections. Adv Exp Med Biol 1050:27–44
Planche T, Aghaizu A, Holliman R, Riley P, Poloniecki J, Breathnach A, Krishna S (2008) Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis 8:777–784
U.S. Food & Drug Administration (2019) Nucleic acid based tests. https://www.fda.gov/medicaldevices/productsandmedicalprocedures/invitrodiagnostics/ucm330711.htm. Accessed 28 Feb 2019
American Society for Microbiology (2010) A practical guidance document for the laboratory detection of toxigenic Clostridium difficile [update of September 9, 2010, version]. September 21, 2010
Bai Y, Sun X, Jin Y, Wang Y, Li J (2017) Accuracy of Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection: a meta analysis. PLoS ONE 12(10):e0185891
McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C (2018) Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48
Whiting PF, Rutjes AW, Westwood ME (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155(8):529–536
Bai YY, Wang YL, Shao CH, Hao YY, Jin Y (2016) Genotype MTBDR plus assay for rapid detection of multidrug resistance in Mycobacterium tuberculosis: a meta analysis. PLoS ONE 11(3):e0150321
Deeks JJ, Macaskill P, Irwig L (2005) The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58:882–893
Huang H, Weintraub A, Fang H, Nord CE (2009) Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of clostridium difficile infections. J Clin Microbiol 47(11):3729–3731
Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P et al (2010) Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol 48(10):3719–3724
Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH et al (2010) Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol 48(3):889–893
Swindells J, Brenwald N, Reading N, Oppenheim B (2010) Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol 48(2):606–608
Goldenberg SD, Dieringer T, French GL (2010) Detection of toxigenic Clostridium difficile in diarrheal stools by rapid real-time polymerase chain reaction. Diagn Microbiol Infect Dis 67(3):304–307
Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Bauer JB, Burnham CAD, Dunne WM Jr (2011) Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol 49(8):2887–2893
Zidarič V, Kevorkijan BK, Oresic N, Janezic S, Rupnik M (2011) Comparison of two commercial molecular tests for the detection of Clostridium difficile in the routine diagnostic laboratory. J Med Microbiol 60(Pt 8):1131–1136
Buchan BW, Mackey TL, Daly JA, Alger G, Denys GA, Peterson LR et al (2012) Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J Clin Microbiol 50(12):3932–6
Viala C, Le Monnier A, Maataoui N, Rousseau C, Collignon A, Poilane I (2012) Comparison of commercial molecular assays for toxigenic Clostridium difficile detection in stools: BD GeneOhm Cdiff, XPert C. difficile and illumigene C. difficile. J Microbiol Methods 90(2):83–5
Shin S, Kim M, Kim M, Lim H, Kim H, Lee K, Chong YS (2012) Evaluation of the Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection. Ann Lab Med 32(5):355–358
Dalpke AH, Hofko M, Zorn M, Zimmermann S (2013) Evaluation of the fully automated BD MAX Cdiff and Xpert C. difficile assays for direct detection of Clostridium difficile in stool specimens. J Clin Microbiol 51(6):1906–8
Eigner U, Fenner I, Veldenzer A, Schwarz R, Oberdorfer K, Holfelder M (2014) Evaluation of six PCR assays in combination with patient related data for the diagnosis of Clostridium difficile-associated infections. Clin Lab 60(8):1343–1350
Gilbreath JJ, Verma P, Abbott AN, Butler-Wu SM (2014) Comparison of the Verigene Clostridium difficile, Simplexa C. difficile Universal Direct, BD MAX Cdiff, and Xpert C. difficile assays for the detection of toxigenic C. difficile. Diagn Microbiol Infect Dis 80(1):13–8
Jensen MB, Olsen KE, Nielsen XC, Hoegh AM, Dessau RB, AtlungEngberg T (2015) Diagnosis of Clostridium difficile: real-time PCR detection of toxin genes in faecal samples is more sensitive compared to toxigenic culture. Eur J Clin Microbiol Infect Dis 34(4):727–736
Jazmati N, Wiegel P, Ličanin B, Plum G (2015) Evaluation of the Qiagen artus C. difficile QS-RGQ Kit for detection of Clostridium difficile toxins A and B in Clinical Stool Specimens. J Clin Microbiol 53(6):1942–4
Yoo J, Lee H, Park KG, Lee GD, Park YG, Park YJ (2015) Evaluation of 3 automated real-time PCR (Xpert C. difficile assay, BD MAX Cdiff, and IMDx C. difficile for Abbott m2000 assay) for detecting Clostridium difficile toxin gene compared to toxigenic culture in stool specimens. Diagn Microbiol Infect Dis 83(1):7–10
Moon HW, Kim HN, Kim JY, Hur M, Kim H, Yun YM (2016) Performance of the artus C. difficile QS-RGQ Kit for the detection of toxigenic Clostridium difficile. Clin Biochem 22. pii: S0009–9120(16)30205–3
Moon HW, Kim HN, Hur M, Shim HS, Kim H, Yun YM (2016) Comparison of diagnostic algorithms for detecting toxigenic clostridium difficile in routine practice at a rertiary referral hospital in Korea. PLoS One 11(8):e0161139
Shin BM, Yoo SM, Shin WC (2016) Evaluation of Xpert C. difficile, BD MAX Cdiff, IMDx C. difficile for Abbottm 2000, and Illumigene C. difficile assays for direct detection of toxigenic clostridium difficile in stool specimens. Ann Lab Med 36(2):131–7
Rajabally N, Kullin B, Ebrahim K, Brock T, Weintraub A, Whitelaw A, Bamford C, Watermeyer G, Thomson S, Abratt V, Reid S (2016) A comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African centre. J Med Microbiol 65(4):320–327
Paitan Y, Miller-Roll T, Adler A (2017) Comparative performance study of six commercial molecular assays for rapid detection of toxigenic Clostridium difficile. Clin Microbiol Infect 23:567e572
Seo JY, Jeong JH, Kim KH, Ahn J-Y, Park P-W, Seo Y-H (2017) Laboratory diagnosis of clostridium difficile infection: comparison of Techlab C. Diff Quik Chek Complete, Xpert C. Difficile, and Multistep Algorithmic Approach. J Clin Lab Anal 31(6):e22135
Huang Xu, Cheng cheng L, Ting X, Chen-jie T, Wen zing X, Fang N, Fang W, Cen ya L (2017) Clinical application evaluation of Xpert detection system of Clostridium difficile. Chin J Clin Lab Sci (Chinese) 35(1):36–38
Shi Wu, Fenfen Z, Ni Yuxing Hu, Yunjian CY, Yunsong Yu, Haihui H (2019) Applicability of commercial multiplex kit for the diagnosis of: a multicenter investigation Clostridium difficile infections. Chin J Lab Med (Chinese) 42(8):674–678
Tenover FC, Baron EJ, Peterson LR, Persing DH (2011) Laboratory diagnosis of Clostridium difficile infection can molecular amplification methods move us out of uncertainty? J Mol Diagn 13:573e582
Barbut F, Surgers L, Eckert C, Visseaux B, Cuingnet M, Mesquita C, Pradier N, Thiriez A, Ait-Ammar N, Aifaoui A, Grandsire E, Lalande V (2014) Does a rapid diagnosis of Clostridium difficile infection impact on quality of patient management? Clin Microbiol Infect 20(2):136–144
Sutton AJ, Higgins JP (2008) Recent developments in meta-analysis. Stat Med 27:625–650
Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y (2010) Chapter 10: analysing and presenting results, p 20. In: Deeks JJ, Bossuyt PM, Gatsonis C (eds) Cochrane handbook for systematic reviews of diagnostic test accuracy version 10. The Cochrane Collaboration, London
Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ (2016) European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22(Suppl 4):S63–S81
Burnham CA, Carroll KC (2013) Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 26:604–630
Chow SK, Naderpour A, Van Enk J (2019) It is not about the assay: preanalytical screening is the key to reducing Clostridioides difficile infection. J Clin Microbiol 57(2):e01553-e1618
DaviesID KA, Planche T, Wilcox MH (2018) The predictive value of quantitative nucleic acid amplification detection of Clostridium difficile toxin gene for faecal sample toxin status and patient outcome. PLoS ONE 13(12):e0205941
Crobach MJT, Duszenko N, Terveer EM, Verduin CM, Kuijper EJ (2018) Nucleic acid amplification test quantitation as predictor of toxin presence in Clostridium difficile infection. J Clin Microbiol 56(3):e01316-e1317
Funding
This work was supported by the Natural Science Foundation of Shandong Province (grant number ZR2016HB44).
Author information
Authors and Affiliations
Contributions
BYY and WYL conceived the study and wrote the manuscript. HYY designed the experiments. SZ and CWJ analyzed the data, contributed to preparing the final version of the manuscript, and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Mara Correa Lelles Nogueira
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bai, Y., Hao, Y., Song, Z. et al. Evaluation of the Cepheid Xpert C. difficile diagnostic assay: an update meta-analysis. Braz J Microbiol 52, 1937–1949 (2021). https://doi.org/10.1007/s42770-021-00563-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00563-7