Abstract
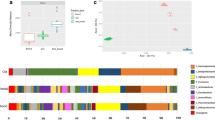
Honey bee colony losses worldwide call for a more in-depth understanding of the pathogenic and mutualistic components of the honey bee microbiota and their relation with the environment. In this descriptive study, we characterized the yeast and bacterial communities that arise from six substrates associated with honey bees: corbicular pollen, beebread, hive debris, intestinal contents, body surface of nurses and forager bees, comparing two different landscapes, Minas Gerais, Brazil and Maryland, United States. The sampling of five hives in Brazil and four in the USA yielded 217 yeast and 284 bacterial isolates. Whereas the yeast community, accounted for 47 species from 29 genera, was dominated in Brazil by Aureobasidium sp. and Candida orthopsilosis, the major yeast recovered from the USA was Debaryomyces hansenii. The bacterial community was more diverse, encompassing 65 species distributed across 31 genera. Overall, most isolates belonged to Firmicutes, genus Bacillus. Among LAB, species from Lactobacillus were the most prevalent. Cluster analysis evidenced high structuration of the microbial communities, with two distinguished microbial groups between Brazil and the United States. In general, the higher difference among sites and substrates were dependents on the turnover effect (~ 93% of the beta diversity), with a more pronounced effect of nestedness (~ 28%) observed from Brazil microbiota change. The relative abundance of yeasts and bacteria also showed the dissimilarity of the microbial communities between both environments. These results provide a comprehensive view of microorganisms associated with A. mellifera, highlighting the importance of the environment in the establishment of the microbiota associated with honey bees.




Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Berenbaum MR, Liao L-H (2019) Honey bees and environmental stress: toxicologic pathology of a superorganism. Toxicol Pathol. https://doi.org/10.1177/0192623319877154
Hölldobler B, Wilson EO (2008). The superorganism: the beauty, elegance and strangeness of insect societies. WW Norton & Company
Lamei S, Stephan JG, Nilson B et al (2019) Feeding honeybee colonies with honeybee-specific lactic acid bacteria (Hbs-LAB) does not affect colony-level Hbs-LAB composition or Paenibacillus larvae spore levels, although american foulbrood affected colonies harbor a more diverse Hbs-LAB community. Microb Ecol. https://doi.org/10.1007/s00248-019-01434-3
Hughes DP, Pierce NE, Boomsma JJ (2008) Social insect symbionts: evolution in homeostatic fortresses. Trends Ecol Evol 23:672–677
Khan KA, Al-Ghamdi AA, Ghramh HA, Ansari MJ et al (2020) Structural diversity and functional variability of gut microbial communities associated with honey bees. Microb Pathog 138:103793. https://doi.org/10.1016/j.micpath.2019.103793
Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffman G (2011) An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis mellifera). Insect Soc 58:431–444
Vásquez A, Forsgen E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC (2012) Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS ONE 7(3):e33188
Gilliam M (1979) Microbiology of pollen and bee bread: the yeasts. Apidologie 10:43–53
Rosa CA, Lachance MA (1998) The yeast genus Starmerella gen. nov. and Starmerella bombicola sp. nov., the teleomorph of Candida bombicola (Spencer, Gorin & Tullock) Meyer & Yarrow. Int J Syst Bacteriol 48:1413–1417
Rosa CA, Lachance MA, Silva JOC, Teixeira ACP, Marini MM, Antonini Y, Martins RP (2003) Yeast communities associated with stingless bees. FEMS Yeast Res 4:271–275
Daniel HM, Rosa CA, São Thiago-Calaça PS, Antonini Y, Bastos EM, Evrard P, Huret S, Fidalgo-Jiménez A, Lachance MA (2013) Starmerella neotropicalis f.a., sp. nov., a yeast species found in bees and pollen. Int J Syst Evol Microbiol 63:3896–3903
Paludo CR, Menezes C, Silva-Junior EA, Vollet-Neto A et al (2018) Stingless bee larvae require fungal steroid to pupate. Sci Rep 8:1122. https://doi.org/10.1038/s41598-018-19583-9
Matos TTS, Teixeira JF, Macías LG, Santos ARO, Suh SO, Barrio E, Lachance MA, Rosa CA (2020) Kluyveromyces osmophilus is not a synonym of Zygosaccharomyces mellis; reinstatement as Zygosaccharomyces osmophilus comb. nov. Int J Syst Evol Microbiol 70:3374–3378. https://doi.org/10.1099/ijsem.0.004182
Gilliam M (1979) Microbiology of pollen and bee bread: the genus Bacillus. Apidologie 10:269–274
Gilliam M (1997) Identification and roles of non-pathogenic microflora associated with honey bess. FEMS Microbiol Lett 155:1–10
Anderson KE, Sheehan TH, Mott BM, Maes P, Snyder L et al (2013) Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 8(12):e83125
Aizenberg-Gershtein Y, Izhaki I, Halpern M (2013) Do honeybees shape the bacterial community composition in floral nectar? PLoS ONE 8(7):e67556
Kwong WK, Moran N (2016) Gut microbial communities of social bees. Nature Reviews: Microbiology. https://doi.org/10.1038/nrmicro.2016.43
Evans JD, Spivak M (2010) Socialized medicine: Individual and communal disease barriers in honey bees. J Invertebr Pathol 103:S62–S72
Human H, Nicolson SW (2006) Nutritional content of fresh, bee collected and stored pollen of Aloe greatheadii var. davyana (Asphodelaceae). Phytochem 67:1486–1492
Carroll MJ, Brown N, Goodall C, Downs AM, Sheenan TH, Anderson KE (2017) Honey bees preferentially consume freshly-stored pollen. PLoS ONE 12(4):e0175933. https://doi.org/10.1371/journal.pone.0175933
Forsgren E, Laugen AT (2014) Prognostic value of using bee and hive debris samples for the detection of American foulbrood disease in honey bee colonies. Apidologie 45:10–20
Starmer WT, Lachance MA (2011) Yeast ecology. In: Kurtzman CP, Fell J, Boekhout T (eds) The yeasts: a taxonomic study, 5th edn. Elsevier
MaddenAA EMJ, Fukami T, Irwin RE, Sheppard J, Sorger DM, Dunn RR (2018) The ecology of insect–yeast relationships and its relevance to human industry. Proc R Soc B 285:20172733. https://doi.org/10.1098/rspb.2017.2733
Jones JC, Fruciano C, Hildebrand F, Toufalilia HA, Balfour NJ et al (2017) Gut microbiota composition is associated with environmental landscape in honey bees. Ecol Evol 8:441–451
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The Yeasts, a Taxonomic Study, 5th edn. Elsevier Science, Amsterdam, pp 87–110
Sambrook J, Fritsch EF, Maniatis T (1986) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Habor Laboratory Press, Cold Spring Habor
Lieckfeldt E, Meyer W, Borner T (1993) Rapid identification and differentiation of yeast by DNA and PCR fingerprinting. J Basic Microb 33:413–426
Silva-Filho EA, Santos SKB, Resende AM, Morais JOF, Morais-Jr MA, Simões DA (2005) Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting. Antonie Van Leeuwenhoek 88:13–23
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Lachance MA, Bowles JM, Starmer WT, Barker JSF (1999) Kodamaea kakaduensis and Candida tolerans, two new yeast species from Australian Hibiscus flowers. Can J Microbiol 45:172–177
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA 6: Molecular Evolutionaty Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 21:403–410
Hoffman CS, Winstonm F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Baselga A (2017) Partitioning abundance-based multiple-site dissimilarity into components: balanced variation in abundance and abundance gradients. Methods Ecol Evol 8:799–808
Baselga A (2010) Partitioning the turnover and nestedness components of beta diversity. Global Ecol Biogeogr 19:134143
Baselga A (2013) Separating the two components of abundance-based dissimilarity: balanced changes in abundance vs. abundance gradients. Methods Ecol Evol 4:552–557
Johnson BR (2008) Within-nest temporal polyethism in the honey bee. Behav Ecol Sociobiol 62:777–784. https://doi.org/10.1007/s00265-007-0503-2
Yun JH, Jung MJ, Kim PS, Bae JW (2018) Social status shapes the bacterial and fungal gut communities of the honey bee. Scientific Reports. https://doi.org/10.1038/s41598-018-19860-7
Gildemacher P, Heijne B, Silvestri M, Houbraken J, Hoekstra E, Boekhout T (2006) Interactions between yeasts, fungicides and apple fruit russeting. FEMS Yeast Re 6:1149–1156
Lachance MA, Starmer WT, Rosa CA, Bowles JM, Barker JSF, Janzen DH (2001) Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res 1:1–8
Gilliam M, Prest DB, Lorenz BJ (1989) Microbiology of pollen and bee bread: taxonomy and enzymology of molds. Apidologie 20:53–68
Villegas-Plaza M, Figueroa-Ramírez J, Portillo C, Monserrate P, Tibata V, Sanchez HJ (2018) Yeast and bacterial composition in pot-pollen recovered from Meliponini in Colombia: prospects for a promising biological resource. In: Vit P, Pedro SRM, Roubik DW (eds) Pot-Pollen in Stingless Bee Melittology. Springer, New York, pp 263–279
Phaff HJ, Starmer WT (1987) Yeasts associated with plants, insects and soil. In: Rose AH, Harrison JS (eds) The yeasts, vol 1. Biology of Yeasts. Academic Press, London, pp 123–180
Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman, CP (2011) Candida Berkhout (1923). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, taxonomic study, 5th edn, vol. 2. Amsterdam: Elsevier, pp 987-1278
Liu X, Yi Z, Ren Y, Li Y, Hui F (2016) Five novel species in the Lodderomyces clade associated with insects. Int J Syst Evol Microbiol 66:4881–4889
Opulente DA, Langdon QK, Buh KV, Haase MAB, Sylvester K et al (2019) Pathogenic budding yeasts isolated outside of clinical settings. FEMS Yeast Research 19:foz032
Sampaio JP (2011a) Rhodotorula Harrison (1928): Rhodotorula acheniorum (Buhagiar & J.A. Barnett) Rodrigues de Miranda (1975). In: Kurtzman CP, Fell JW, Boekhout T (eds) The Yeasts, a Taxonomic Study, 5ª ed., Amsterdam: Elsevier
Sampaio JP (2011b) Rhodotorula Harrison (1928): Rhodotorula mucilaginosa (Jorgensen) F.C. Harrison (1928). In: Kurtzman CP, Fell JW, Boekhout T (eds). The Yeasts, a Taxonomic Study, 5ª ed., Amsterdam: Elsevier Science, pp 1873-1928
Barriga EJC, Barahona PP, Tufiño C, Bastidas B, Guamán-Burneo C, Freitas L, Rosa CA (2014) An overview of the yeast biodiversity in the Galápagos islands and other ecuadorian regions. In: Grillo O (ed) Biodiversity - The Dynamic Balance of the Planet, IntechOpen, https://doi.org/10.5772/58303
Pozo MI, Herrera CM, Bazaga P (2011) Species richness of yeast communities in floral nectar of Southern Spanish plants. Microb Ecol 61:82–91
Pozo MI, Bartlewicz J, Oystaeyen A, Benavente A, Kemenade G, Wäckers F, Jacquemyn H (2018) Surviving in the absence of flowers: do nectar yeasts rely on overwintering bumblebee queens to complete their annual life cycle? FEMS Microbiol Ecol 1:94. https://doi.org/10.1093/femsec/fiy196
Ergin Ç, Ilkit M, Kaftanoglu O (2004) Detection of Cryptococcus neoformans var. grubii in honeybee (Apis mellifera) colonies. Mycoses 47:431–434
Randhawa HS, Kowshik T, Chowdhary A, Prakash A, Khan ZU, Xu J (2011) Seasonal variations in the prevalence of Cryptococcus neoformans var. grubii and Cryptococcus gattii in decayed wood inside trunk hollows of diverse tree species in north-western India: a retrospective study. Med Mycol 49:320–323
Castro e Silva DM, Santos DCS, Martins MA, Oliveira L, Szeszs MW, Melhem MSC (2015) First isolation of Cryptococcus neoformans genotype VNI MAT-alpha from wood inside hollow trunks of Hymenaea courbaril. Medical Mycology 54:97-102
Santos ARO, Leon MP, Barros KO, Freitas LFD, Hughes AFS, Morais PB, Lachance MA, Rosa CA (2018) Starmerella camargoi f.a., sp. nov., Starmerella ilheusensis f.a., sp. nov., Starmerella litoralis f.a., sp. nov., Starmerella opuntiae f.a., sp. nov., Starmerella roubikii f.a., sp. nov. and Starmerella vitae f.a., sp. nov., isolated from flowers and bees, and transfer of related Candida species to the genus Starmerella as new combinations. Int J Syst Evol Microbiol 68:1333–1343. https://doi.org/10.1099/ijsem.0.002675
Dharampal PS, Diaz-Garcia L, Haase MAB, Zalapa J, Currie CR, Hittinger CT, Steffan SA (2020) Microbial diversity associated with the pollen stores of captive-bred bumble bee colonies. Insects 11:250. https://doi.org/10.3390/insects11040250
Detry R, Simon-Delso N, Bruneau E, Daniel H-M (2020) Specialisation of yeast genera in different phases of bee bread maturation. Microorganisms 8:1789. https://doi.org/10.3390/microorganisms8111789
Costa-Neto DJ, Morais PB (2020) The vectoring of Starmerella species and other yeasts by stingless bees in a Neotropical savanna. Fungal Ecol 47:100973. https://doi.org/10.1016/j.funeco.2020.100973
Fonseca Á, Inácio J (2006) Phylloplane yeasts. In: Péter G, Rosa CA (eds) Biodiversity and ecophysiology of yeasts. Springer, Berlin, pp 263–301
Calaça PSST (2011) Aspectos da biologia de Melipona quinquefasciata Lepeletier (Mandaçaia do chão), características físico-químicas do mel, recursos alimentares e leveduras associadas. Dissertation, Universidade Federal de Ouro Preto
Snowdon JA, Cliver D (1996) Microorganisms in honey. Int J Food Microbiol 31:1–26
Martorell P, Stratford M, Steels H, Fernández-Espinar MT, Querol A (2007) Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int J Food Microbiol 114:234–242
Sudhadham M, Sihanonth P, Sivichai S, Chaiyarat R, Dorrestein GM, Menken SBJ, Hoog GS (2008) The neurotropic black yeast Exophiala dermatitidis has a possible origin in the tropical rain forest. Stud Mycol 61:145–155
Limtong S, Nasanit R (2017) Phylloplane yeasts in tropical climates. In: Buzzini P, Lachance MA, Yurkov A (eds) Yeasts in Natural Ecosystems: Diversity. Springer, Cham
Landell M (2006) Biodiversidade e potencial biotecnológico de leveduras e fungos leveduriformes associados ao filoplano de bromélias no Parque de Itapuã-Viamão/RS. Dissertation, Faculdade de Agronomia, Universidade Federal do Rio Grande do Sul
Morais PB, Sousa FMP, Rosa CA (2020) Yeast in plant phytotelmata: Is there a “core” community in different localities of rupestrian savannas of Brazil? Braz J Microbiol. https://doi.org/10.1007/s42770-020-00286-1
Belisle M, Peay KG, Fukami T (2012) Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microbiol Ecol 63:711–718
Melo WGP (2009) Distribuição biogeográfica de leveduras associadas a exsudados de Calophyllum brasiliense (CLUSIACEAE), em áreas de ecótono da planície do Araguaia – TO. Dissertation, Universidade Federal do Tocantins
Silva MS, Rabadzhiev Y, Eller MR, Iliev I, Ivanova I, Santana WC (2017) Microorganisms in Honey. In: Arnaut De Toledo V (ed) Rijeka Honey Analysis. IntechOpen, https://doi.org/10.5772/63259
Lachance MA, Bowles JM, Díaz MCC, Janzen DH (2001) Candida cleridarum, Candida tilneyi and Candida powelli, three new yeast species isolated from insects associated with flowers. Int J Syst Evol Microbiol 51:1201–1207
McFrederick QS, Rehan SM (2018) Wild bee pollen usage and microbial communities co-vary across landscapes. Microbial Ecol 77:513–522. https://doi.org/10.1007/s00248-018-1232-y
Ganter PF (2006) Yeast and invertebrate associations ecosystems. In: Rosa C, Péter G (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlim, pp 303–370
Tauber JP, Nguyen V, Lopez D, Evans JD (2019) Effects of a resident yeast from the honeybee gut on immunity, microbiota, and nosema disease. Insects 10:296. https://doi.org/10.3390/insects10090296
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92
Moran NA, Hansen AK, Powell JE, Sabree ZL (2012) Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 7:e36393. https://doi.org/10.1371/journal.pone.0036393
Zheng H, Steele MI, Leonard SP, Motta EVS, Moran NA (2018) Honey bees as models for gut microbiota research. Lab Anim 47:317–325. https://doi.org/10.1038/s41684-018-0173-x
Ellegaard KM, Engel P (2019) Genomic diversity landscape of the honey bee gut microbiota. Nat Commun 10:446. https://doi.org/10.1038/s41467-019-08303-0
Kaltenpoth M (2009) Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol 17:12. https://doi.org/10.1016/j.tim.2009.09.006
Fridman S, Izhaki I, Gerchman Y, Halpern M (2012) Bacterial communities in floral nectar. Environ Microbiol Rep 4:97–104
Olofsson TC, Vàsquez A (2008) Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol 57:356363
Bottacini F, Milani C, Turroni F, Sánchez B, Foroni E, Duranti S et al (2012) Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS ONE 7:e44229. https://doi.org/10.1371/journal.pone.0044229
Corby-Harris V, Maes P, Anderson KE (2014) The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 9:e95056
Olofsson TC, Butler È, Markowicz P, Lindholm C, Larsson L, Vásquez A (2014) Lactic acid bacterial symbionts in honey bees, an unknown key to honey’s antimicrobial and therapeutic activities. Int Wound J 13:668–679. https://doi.org/10.1111/iwj.12345
Evans JD, Armstrong TN (2006) Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecol 6:64. https://doi.org/10.1186/1472-6785-6-4
Bavykin SG, Yuri P, Lysov YPL, Zakhariev V, Kelly JJ, Jackman J, Stahl DA, Cherni A (2004) Use of 16S rRNA, 23S rRNA, and gyrB gene sequence analysis to determine phylogenetic relationships of Bacillus cereus group microorganisms. J Clin Microbiol 42:3711–3730
Cano RJ, Borucki MK, Higby-Schweitzer M, Poinar HN, Poinar JRGO, Pollard KJ (1994) Bacillus DNA in fossil bees: an ancient symbiosis? Appl Environ Microbiol 60:2164–2167
Alippi AM, Reynaldi FJ (2006) Inhibition of the growth of Paenibacillus larvae, the causal agent of American foulbrood of honeybees, by selected strains of aerobic spore-forming bacteria isolated from apiarian sources. J Invertebr Pathol 91:141–146
Benitez LB, Velho RV, Motta AS, Segalin J, Brandelli A (2012) Antimicrobial factor from Bacillus amyloliquefaciens inhibits Paenibacillus larvae, the causative agent of American foulbrood. Arch Microbiol 194:177–185. https://doi.org/10.1007/s00203-011-0743-4
Sabaté DC, Carrillo L, Audisio M.C (2009) Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res Microbiol 160 193e199
Endo A, Maeno S, Tanizawa Y, Kneifel W, Arita M, Dicks L, Salminen S (2018) Fructophilic lactic acid bacteria, a unique group of fructose fermenting microbes. Appl Environ Microbiol 84:e01290-18. https://doi.org/10.1128/AEM.01290-18
Vojvodic S, Rehan SM, Anderson KE (2013) Microbial gut diversity of africanized and european honey bee larval instars. PLoS ONE 8:e72106
Endo A, Salminen S (2013) Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst Appl Microbiol 36:444–448
Killer J, Dubná S, Sedláček I, Švec P (2014) Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int J Syst Evol Microbiol 64:152–157. https://doi.org/10.1099/ijs.0.053033-0
Genersch E, Forsgren E, Pentikäinen J, Ashiralieva A, Rauch S, Kilwinski J, Fries I (2006) Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int J Syst Evol Microbiol 56:501–511
Forsgren E (2010) European foulbrood in honey bees. J Invertebr Pathol 103:S5–S9
Jojima Y, Mihara Y, Suzuki S, Yokozeki K, Yamanaka S, Fudou R (2004) Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int J Syst Evol Microbiol 54:2263–2267. https://doi.org/10.1099/ijs.0.02911-0
Baruzzi F, Cefola M, Carito A, Vanadia S, Calabrese C (2012) Changes in bacterial composition of zucchini flowers exposed to refrigeration temperatures. Sci World J, ID 127805https://doi.org/10.1100/2012/127805
Deletóile A, Decré D, Courant S, Passet V, Audo J, Grimont P, Arlet G, Brisse S (2009) Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol 47:300–310
Jeyaprakash A, Hoy MA, Allsopp MH (2003) Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J Invertebr Pathol 84:96–103
Babendreier D, Joller Romeis J, Bigler F, Widmer F (2007) Bacterial community structures in honey bee intestines and their response to two insecticidal proteins. FEMS Microbiol Ecol 59:600–610c
Mohr KI, Tebbe CC (2006) Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol 8:258–272
Martinson VG, Moy J, Moran NA (2011) Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840
Ribière C, Hegarty C, Stephenson H, Whelan P, O’Toole PW (2018) Gut and whole-body microbiota of the honey bee separate thriving and non-thriving hives. Microbial Ecol. https://doi.org/10.1007/s00248-018-1287-9
Qian H, Ricklefs RE, White PS (2005) Beta diversity of Angiosperms angiosperms in temperate floras of eastern Asia and eastern North America. Ecol Lett 8:15–22
Ulrich W, Soliveres S, Maestre FT et al (2014) Climate and soil attributes determine plant species turnover in global drylands. J Biogeogr 41:2307–2319. https://doi.org/10.1111/jbi.12377
Gaston KJ, Blackburn TM (2000) Pattern and process in macroecology. Blackwell Science, Oxford
Subotic S, Boddicker AM, Nguyen VM, Rivers J, Briles CE, Mosier AC (2019) Honey bee microbiome associated with different hive and sample types over a honey production season. PLoS ONE 14(11):e0223834. https://doi.org/10.1371/journal.pone.0223834
Dong Z, Li H, Chen Y, Wang F, Deng X, Lin L, Zhang Q, Li J, Guo J (2020) Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages. Microbiol Res 231:126370
Bonilla-Rosso G, Engel P (2018) Functional roles and metabolic niches in the honey bee gut microbiota. Curr Opin Microbiol 43:69–76
Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan P-L, Briese T, Hornig M, Geiser DM, Martinson V, Engelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin I (2007) A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318:283–287
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Fundação Ezequiel Dias (FUNED).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation and field expeditions were performed by Denise de Oliveira Scoaris and Milton Adolfo Silveira. Data collection and analysis were performed by Denise de Oliveira Scoaris and Frederic Mendes Hughes, under the supervision of Jay Daniel Evans and Carlos Augusto Rosa. The first draft of the manuscript was written by Denise de Oliveira Scoaris and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Melissa Fontes Landell
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Oliveira Scoaris, D., Hughes, F.M., Silveira, M.A. et al. Microbial communities associated with honey bees in Brazil and in the United States. Braz J Microbiol 52, 2097–2115 (2021). https://doi.org/10.1007/s42770-021-00539-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00539-7