Abstract
DNA fulfills two critical roles in a cell by being the template for both DNA replication and RNA transcription. During evolution, eukaryotic cells have adopted multiple strategies to coordinate these two processes to prevent or minimize conflicts that might arise between them. Here, we review the strategies employed by cells to simultaneously accommodate the machineries carrying out DNA replication and transcription, and the mechanisms that are brought into play for resolving conflicts when they do arise. We focus on a group of the so-called ‘difficult-to-replicate’ loci in the human genome, which include chromosome fragile sites, the ribosomal DNA, and telomeres. A failure to resolve conflicts arising between replication and transcription can lead to genome instability, as well as to the initiation of cancer and other age-associated diseases. Understanding the mechanisms required for the resolution of these conflicts could, therefore, open up new therapeutic avenues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deoxyribonucleic acid (DNA) is the macromolecule that stores the genetic information in nearly all living organisms. It is composed of two antiparallel strands coiled around each other in the form of a right-handed helix. For cells to transmit this genetic information to their progeny, DNA needs to be accurately duplicated during the S phase of the cell division cycle, and the resulting sister chromatids must be segregated faithfully to the daughter cells in mitosis. However, the same DNA template is also a substrate for the transcription machinery to make the various different forms of RNA. Transcription has the potential to interfere with DNA replication, because some genes in human cells are so large that their transcription can take a whole cell cycle to complete during which time replication of the same locus has to occur. Moreover, some genes, such as those encoding ribosomal RNAs, must be continuously transcribed to provide a ready supply of ribosomes for protein synthesis. As a result, some regions of the genome have to conduct DNA replication and transcription at the same time, inevitably increasing the probability of a damaging conflict.
DNA replication is initiated at multiple replication origins in eukaryotic cells. During the G1 phase, replication origins are licensed through the recruitment of the origin recognition complex (ORC), which, in turn, recruits Cdc6 and Cdt1, and, subsequently, the non-activated form of the replicative DNA helicase, MCM2–7, to form the so-called pre-replication complex (pre-RC) (Boos et al. 2013). Each licensed origin contains a head-to-head assembled, double-hexameric MCM2–7 complex. Once cells have entered S phase, replication firing factors, including Cdc45, the GINS complex, and MCM2–7 to establish the Cdc45∙MCM2–7∙GINS (CMG) complex (the CMG helicase). Other replication factors (including Mcm10 and Ctf4) are known to facilitate the activation of the CMG helicase and the recruitment of the replicative DNA polymerases (Thu and Bielinsky 2013; Zhu et al. 2007). The CMG complex then initiates DNA unwinding to trigger DNA synthesis (Bleichert et al. 2017). Once a DNA origin is opened to create a replication bubble, two replication forks are established that travel in opposite directions as replication proceeds.
In common with replication, transcription begins at specific genomic regions. To initiate transcription in eukaryotic cells, general transcription factors first bind to a gene promoter, which assists in recruitment of RNA polymerase (RNAP). RNAP must be activated to catalyze the process of transcription through both transcription factor II Human (TFIIH)-mediated DNA denaturation and RNAP phosphorylation. Together with numerous transcription elongation and RNA processing factors, RNAP then catalyzes transcription elongation (Bentley 2014). Unlike replication, transcription does not occur in isolation, and it is always coupled with downstream events required for RNA processing (e.g. gene splicing). Therefore, the large RNAP complex and its associated RNA processing factors present a real challenge for any advancing replication fork that might be encountered.
Since DNA replication and transcription can occur on the same stretch of template DNA at the same time, the multi-enzyme machineries required to catalyze these two processes have the potential to collide with each other. While this might appear to be a catastrophic event for a cell, in most cases, cells can proliferate efficiently despite this potential threat. Therefore, cells must possess defined strategies to orchestrate these two processes to prevent or minimize collisions. Nevertheless, mounting evidence indicates that replication and transcription collisions do occur within certain genomic loci, and that these conflicts are a natural source of genomic instability (Garcia-Muse and Aguilera 2016; Lin and Pasero 2012). In this review, we discuss the strategies employed by cells to coordinate replication and transcription and to limit damaging collisions. We will also discuss the pathways involved in resolving replication-transcription collisions that do occur in the human genome, particularly those arising at specific loci that are deemed to be ‘difficult-to-replicate’, such as fragile sites, the ribosomal DNA (rDNA) genes, and telomeres.
Strategies for coordinating replication and transcription
In bacteria, collisions between the replication and transcription machineries are unavoidable, because the genome is being replicated and transcribed constantly during favorable growth conditions. Gene transcription in bacteria such as E. coli is biased in the genome, such that most genes are transcribed and replicated in the same direction (to avoid head-on collisions). Nevertheless, the replisome travels on the DNA template much faster than does RNAP, and therefore there are inevitable co-directional collisions where the replisome encounters the transcription machinery from behind (Helmrich et al. 2013). In the case of eukaryotic cells, where the genome is much larger, and both replication and transcription can start at multiple loci simultaneously, strategies have evolved to coordinate DNA replication and transcription in a more sophisticated manner to avoid collisions between these two processes.
Spatial separation of replication and transcription
About 20 years ago, analyses in mouse and human fibroblasts showed that sites of DNA replication and transcription are organized within distinct territories throughout the whole period of S phase. This suggests that replication and transcription are spatially separated to avoid the machineries occupying the same section of the genomic template at the same time (Wei et al. 1998) (Fig. 1a). Moreover, transcription-dependent gene looping has the potential to insulate transcription machineries from replication forks, thus limiting collisions (Bermejo et al. 2007). Consistent with this proposal, DNA topoisomerase II is essential both for gene looping and for avoiding replication and transcription collisions (Bermejo et al. 2012; Li et al. 1999).
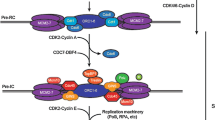
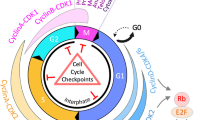
Strategies employed by eukaryotic cells to coordinate replication and transcription. a Sites of DNA replication and transcription occur in separate domains in either early or late S phase, particularly in the nucleolar area where the rDNA loci reside. b Timing of transcription and replication is highly regulated. Most of the transcription occurs before S phase and fades in S phase. In mammalian cells, the transition from G1 to S phase is controlled by the Rb/E2F complex. In the absence of Rb, E2F1 binds to the promoters of target genes encoding proteins involved in DNA replication (e.g., DNA polymerases, thymidine kinase, dihydrofolate reductase, and CDC6), and chromosomal replication (e.g., replication origin-binding proteins ORC1 and MCM5). As a result, it facilitates the G1/S transition and S-phase progress. When cells are not proliferating, E2F is bound to Rb protein contributing transcriptional suppression. c In eukaryotic cells, there is a bias towards co-orientation of replication and transcription. Top panel, in areas where highly transcribed or large genes reside, replication initiation sites frequently co-localize with transcription regulatory sites including enhancers or promoters. Bottom panel, in the regions where genes are highly transcribed, i.e., rDNA regions, sections of specific DNA sequences are bound by proteins (e.g., Fob1 or Sap1 in yeasts) that can serve as barriers for the replication fork coming from the opposite direction to that of transcription
Temporal separation of replication and transcription
It has been demonstrated that more than 95% of the sites of ongoing replication do not overlap with sites of transcription in early S phase (Wei et al. 1998), suggesting that replication and transcription can also be separated in a temporal manner (Fig. 1b). Indeed, in the heavily transcribed rDNA regions where transcription and replication cannot be easily divided spatially, replication timing varies in the different units of these highly repetitive regions. For example, it has been shown that rDNA regions containing actively transcribed units replicate in early S phase, while those regions that do not have active units replicate late in S phase (Dimitrova 2011). More generally, nascent RNA sequencing analysis has confirmed that the timing of DNA replication and that of transcription differ globally in human cells (Meryet-Figuiere et al. 2014). The molecular mechanisms underlying the regulation of replication timing remain to be fully elucidated, although several advances have been made in the past decade. For example, the high-resolution chromosome conformation capture technique has revealed that the formation of topologically associated domains (clusters of self-interacting regions) is vital for minimizing replication and transcription conflicts (Marchal et al. 2019; Pope et al. 2014).
It would be predicted from the above discussion that an alteration in replication timing would provoke collisions between the replication and transcription machineries leading to genome instability (Almeida et al. 2018; Herlihy and de Bruin 2017). Indeed, many proteins identified as playing a role in controlling replication timing (either locally or globally) are also involved in the cellular response to DNA damage generated during S phase (Yamazaki et al. 2013). For example, in fission yeast, the telomere-binding protein, Taz1, binds to late replication regions to regulate replication timing (Tazumi et al. 2012), and is required for the prevention and repair of DNA breaks at these loci (Foti et al. 2016). In budding yeast, Fhk1 and Fhk2 control replication initiation timing globally through influencing origin clustering (Knott et al. 2012), and their function is closely related to cell-cycle regulation, cellular stress responses, and aging (Murakami et al. 2010). In mammal cells, RIF1 is an important, non-replisome-associated, protein that plays a genome-wide role in regulation of replication-timing (Yamazaki et al. 2013). This is apparently achieved through its combined ability to modulate the interaction between different replicating domains in G1 cells, and to bind to late-replicating regions to prevent unscheduled origin firing (Cornacchia et al. 2012; Foti et al. 2016; Yamazaki et al. 2012).
Besides replication timing, transcription patterns are also strictly regulated throughout the cell cycle, especially in readiness for the transition from G1 to S phase (Wittenberg and Reed 2005). It is well established that transcription of genes required for S phase is tightly controlled by the E2F family of transcription factors (Dimova and Dyson 2005). This wave of transcription starts in early G1 phase, peaks at the G1/S boundary, and then fades during the early stages of S phase (Bertoli et al. 2013). One of the proposed biological reasons behind this is to prevent the interference between replication and transcription (Fig. 1b). Consistent with this idea, overexpression of oncogenes, such as those encoding MYC or Cyclin E, can lead to increased E2F activity and premature S phase entry. As a consequence, the global G1 gene transcription program fails to be completed before S phase entry, leading to a high level of transcription throughout S phase (Bertoli et al. 2016). Through examining nascent DNA synthesis in early S phase under conditions of Cyclin E or MYC overexpression in human cells, unscheduled DNA synthesis from origins located in intragenic regions has been observed (Macheret and Halazonetis 2018). These intragenic origins are normally disabled by transcription during G1 (Macheret and Halazonetis 2018), which suggests that their inappropriate firing was caused by premature S phase entry, because transcription did not have sufficient time to displace the pre-replicative complexes from these origins. As a result, conflicts between replication and transcription would occur when these origins fire, potentially leading to DNA double-strand break formation and the subsequent chromosomal rearrangements (Macheret and Halazonetis 2018).
Co-orientation of replication and transcription
In a situation where replication and transcription must occur at the same time in a particular genomic locus, cells try to avoid conflicts between these two machineries by creating a bias in favor of co-directional movement of the replication and transcription machineries. In bacterial cells, it is well established that highly transcribed rRNA, tRNA, and other essential genes are almost exclusively co-directional with replication fork progression (Guy and Roten 2004; Rocha and Danchin 2003). In eukaryotic cells, several features of genome organization are utilized to provide a similar co-directional bias. Replication initiation sites are frequently found within transcription regulatory units, including enhancers and promoters. For example, replication origins in S. cerevisiae are recognized by transcription factors (Herrgård et al. 2006; Miotto et al. 2016). In mammals, although no such consensus elements are evident, replication origins tend to overlap with enhancers and promoters (Cadoret et al. 2008; Miotto et al. 2016; Sequeira-Mendes et al. 2009). Recently, cis elements involved in controlling replication timing have been identified in mouse embryonic stem cells (Sima et al. 2019). These elements often possess the features of enhancers or promoters, enforcing the notion that replication and transcription are coupled. In addition, numerous lines of evidence indicate that transcriptional initiation activity defines those replication origins that will fire efficiently (Candelli et al. 2018; Chen et al. 2019; Sequeira-Mendes et al. 2009). This, in turn, enables replication and transcription to occur in a co-directional fashion. Consistent with this, using an Okazaki fragment sequencing technique in human RPE-1 cells, Chen et al. showed that replication origins preferentially fire near RNAPII-occupied transcription start sites of highly transcribed genes, creating a strong bias towards co-directional replication and transcription (Chen et al. 2019) (Fig. 1c).
Another strategy to impose bias is to use the so-called ‘replication fork barriers’, which are found in many eukaryotic cells, and enable co-directional replication and transcription at the highly transcribed rDNA loci (Gerber et al. 1997; Little et al. 1993). These replication fork barriers are composed of defined DNA sequences and are tightly bound by non-nucleosomal proteins (e.g., Fob1 in S. cerevisiae and Sap1 in Schizosaccharomyces pombe). They are located at the 3′ termini of rDNA transcriptional units to ensure that any replication forks progressing in the opposite direction to the direction of transcription will be stalled before colliding with RNA polymerase I (Muller et al. 2000; Pasero et al. 2002) (Fig. 1c). Similar pausing sites have been found at tRNA regions in yeast, and are required to stop replication forks only if they are progressing in the opposite direction to that of tRNA transcription (Deshpande and Newlon 1996; Ivessa et al. 2003).
Consequences of conflicts between replication and transcription
Despite the strategies adopted by cells to prevent the DNA replication and transcription machineries colliding with each other, it is inevitable that such collisions occur occasionally. It is generally believed that co-directional encounters are less detrimental to cells. Indeed, in vitro, it was shown that a reconstituted E. coli replisome can displace or bypass a co-directional RNAP, as well as to use the RNA as a primer to conduct DNA synthesis under some circumstances (Liu et al. 1993; Pomerantz and O’Donnell 2008). Whether this mechanism exists in living bacteria (or in eukaryotic cells) remains unclear. However, even co-directional encounters are not always benign. For example, using a well-defined episomal replication system in human cells, Hamperl et al. showed that co-directional conflicts promote an ATM-dependent DNA-damage response, suggesting that these conflicts lead to DNA damage (Hamperl et al. 2017).
Understandably, head-on encounters are considered to be more deleterious to fork progression and are widely associated with the creation of DNA damage (Deshpande and Newlon 1996; Merrikh et al. 2012; Prado and Aguilera 2005). The negative consequences of a head-on encounter go beyond those created by a direct collision between the replication and transcription machineries. For example, when the replication and transcription machineries converge, DNA torsional stress in the form of positive supercoiling accumulates in the intervening region, inhibiting helix unwinding and thus preventing continued replication fork progression (Bermejo et al. 2012) (Fig. 2). Indeed, in both yeast and human cells, DNA topoisomerases I and II associate with replication forks, and are required to prevent replication and transcription interference (Bermejo et al. 2012; Tuduri et al. 2009). Head-on encounters can also induce pathological R-loop formation involving the nascent RNA transcript (Fig. 2). R-loops are three-stranded nucleic acid structures containing one DNA–RNA hybrid and a displaced single strand of DNA. These structures can directly block DNA replication, leading to fork stalling or collapse, and can induce a DNA-damage response (Aguilera and García-Muse 2012; Santos-Pereira and Aguilera 2015). Consistent with this, R-loops have been shown to be significantly enriched at sites where head-on encounters are prevalent (Hamperl et al. 2017).
Summary of the pathways involved in preventing or resolving replication and transcription conflicts in eukaryotic cells in S phase. When conflicts between replication and transcription are unavoidable, several pathways are available to lessen the impact of or resolve the conflicts. First, the newly synthesized RNAs are coated with RNA processing proteins to prevent R-loop formation. These proteins are referred as messenger ribonucleoprotein complexes (mRNPs, e.g., the THO complex and splicing factors ASF/SF2). Second, R loops can be resolved by either RNaseH or helicases such as SETX (Senataxin) or DHX9. In some cases, the displaced DNA strand might form a G-quadruplex (G4) that would stabilize the R-loop structure. Third, stalled RNAP blocking the replisome can be removed by either the transcription-coupled nucleotide repair pathway (TC-NER), Def1-mediated, proteasome-dependent degradation, or via the Dicer-mediated digestion of the newly synthesized RNA and the subsequent degradation of RNAP. Finally, anti-backing of RNAP facilitated by TFIIS and RECQL5 in human cells may also prevent potential collisions between transcription and replication. With the above measures, the conflicts between replication and transcription could be resolved even in the case of a head-on collision. When, however, the conflicts cannot be resolved, the DNA between the two machineries might become positively supercoiled, while DNA behind the transcription becomes negatively supercoiled, which would increase the chance of R-loop formation and subsequent genome instability
Pathways for resolving replication–transcription conflicts
Fortunately, when a collision between the replication and transcription machineries does occur, cells are equipped with efficient surveillance and repair pathways to deal with the wide range of atypical and/or undesirable structures that might be formed. These include DNA secondary structures, such as G-quadruplexes and hairpins, R-loops, damage to the DNA template (e.g. collapsed replication forks), as well as backtracked or stalled RNAPs. Indeed, genome-wide analyses have indicated that transcribed genes are natural pausing sites for DNA replication (Azvolinsky et al. 2009), indicating that cells must have efficient means to overcome these impediments.
R-loop prevention and resolution
As discussed above, one of the by-products of transcription are R-loops, which are very stable in vitro and in vivo. R-loops can arrest both DNA replication forks and RNA polymerases (Aguilera and García-Muse 2012; Belotserkovskii and Hanawalt 2011; Belotserkovskii et al. 2010, 2017). A current model for how co-transcriptional R-loops are formed is the ‘thread-back’ model, where the newly synthesized RNA can invade the transiently opened template DNA (Aguilera and García-Muse 2012). One of the main strategies to prevent co-transcriptional R-loop formation is the coupling of transcription to mRNA processing and export. The newly synthesized RNA is immediately bound by proteins, including the THO complex and splicing factors ASF/SF2, which prevents the nascent RNA from hybridizing to the template DNA (Aguilera and García-Muse 2012; Huertas and Aguilera 2003; Li and Manley 2005) (Fig. 2). Another strategy is to prevent the accumulation of negative supercoiling behind the advancing RNAP, which would, otherwise, facilitate helix opening and, hence, RNA/DNA annealing. This function is dependent on DNA topoisomerase I, whose role in limiting R-loop formation is well documented (Drolet et al. 1995; El Hage et al. 2010; Tuduri et al. 2009).
Cells have also developed multiple strategies to resolve R-loops and, hence, minimize their deleterious effects. First, the RNA component can be degraded. In yeast and human cells, ribonuclease H1 (RNase H1) and ribonuclease H2 (RNase H2) can specifically degrade the RNA strand of a DNA/RNA hybrid. In human cells, RNase H1 seems more important in the context of transcription-associated R-loops, and has a genome-wide role in resolving R-loops. However, a recent study demonstrated that RNase H2 plays an essential role in R-loop processing and ribonucleotide excision repair in the G2 and M phases in yeast (Lockhart et al. 2019). RNase H1, however, can function independently of the cell cycle to remove R-loops and appears to become activated in response to high R-loop loads in S phase. It would be interesting to determine if this is also the case in human cells. Second, the R-loop can be resolved after it has encountered a replication fork. The fork stalling generated under these circumstances actives the Fanconi anemia–BRCA pathway to remove R-loops, ensuring the complete duplication of genome in S phase (García-Rubio et al. 2015; Madireddy et al. 2016; Schwab et al. 2015). Third, certain helicases have been identified that specifically disassemble R loops at specific loci, including Senataxin (SETX) and DHX9. The RNA/DNA helicase SETX is one of the best characterized R-loop-binding factors. Mutations in the SETX gene have been found in two neurodegenerative disorders: ataxia with oculomotor apraxia type 2 (AOA2) and amyotrophic lateral sclerosis type 4 (ALS4) (Chen et al. 2004; Moreira et al. 2004). It was demonstrated that BRCA1 can recruit SETX to the termination regions of gene transcription where R-loop formation is known to occur naturally, and this prevents the accumulation of single-strand DNA breaks (SSBs) at those regions (Hatchi et al. 2015). Most interestingly, when SETX is conjugated to SUMO2/3, it can resolve transcription–replication encounters by interacting with the 3–5′ exonuclease exosome complex via the EXOSC9/RRP45 subunit (Richard et al. 2013) (Fig. 2).
RNAP complex clearance and prevention of backtracking
In E. coli, the actions of two DNA repair proteins, Mfd and UvrD, can displace a stalled RNAP by pulling the RNAP back from the stall site (Epshtein et al. 2014; Ganesan et al. 2012). Similarly, in eukaryotes, the transcription-coupled nucleotide excision repair pathway helps to remove RNAP (Vermeulen and Fousteri 2013). If such strategies fail, persistent RNAP complexes can be ubiquitylated and degraded through the proteasome-dependent pathway (Wilson et al. 2013) or released using components of the RNA interference pathway (Castel et al. 2014; Zaratiegui et al. 2011) (Fig. 2).
In addition, RNAP oscillates between productive and backtracked states at numerous DNA positions. ‘RNAP backtracking’ refers to the state where RNAP reverses on the template by one or more nucleotides, and is essential for the maintenance of transcriptional fidelity (Artsimovitch and Landick 2000; Nudler 2012). Deep sequencing of 3′ ends of nascent transcripts associated with RNAPII in yeast has indicated that 75% of DNA polymerase pausing sites are associated with RNAP backtracking (Churchman and Weissman 2011). In contrast to this physiological RNAP backtracking, unscheduled backtracking can lead to DNA damage and is proposed to be responsible for the formation of at least some of the double-strand DNA breaks generated during co-directional encounters (Dutta et al. 2011) (Fig. 2). In general, RNAP backtracking events are rarely seen during the elongation phase of transcription. However, when encountering obstacles, such as damaged DNA or stably bound proteins, RNAP backtracking can occur during transcription elongation. In this situation, RNAP may lose contact with the end of the nascent transcript leading to an arrested RNAP that will eventually be encountered by the replication machinery (Gomez-Herreros et al. 2012).
Luckily, anti-backtracking processes exist to prevent any deleterious consequence caused by RNAP backtracking. In prokaryotes, transcription can be directly coupled with translation due to the lack of a nuclear membrane (Proshkin et al. 2010). In addition, some transcription factors, including TFIIS in eukaryotes (Thomas et al. 1998), can assist transcription elongation by reducing nucleotide mis-incorporation, which is the main cause of RNAP backtracking, thereby preventing the replication and transcription collisions. Moreover, backtracked RNAP can be reactivated by specific factors, including GreA and GreB in bacteria (Opalka et al. 2003) and TFIIS in eukaryotes (Cheung and Cramer 2011). These factors can cleave the transcript to produce a new 3′-OH end, such that transcription can resume. Interestingly, human RECQL5 is also proposed to prevent replication and transcription conflicts by restricting RNAP backtracking (Saponaro et al. 2014; Urban et al. 2016) (Fig. 2).
Hotspots of replication and transcription conflicts
Although cells possess numerous mechanisms to prevent or resolve replication and transcription conflicts, certain genomic regions seemingly encounter more conflicts due to the nature of their DNA sequence or genomic location, or their association with transcribed genes and transcription factor binding sites (i.e., enhancers and promoters). As a result, these regions are deemed to be hard-to-replicate and are prone to be unstable.
Fragile sites
Chromosome fragile sites are genomic regions displaying visible breaks or gaps (fragility) on metaphase chromosomes (Sutherland 1979) following replication perturbation (often referred to as replication stress; RS). Based on their frequency in the population, these loci are classified as either common fragile sites (CFSs), which are present in all individuals, or rare fragile sites (RFSs), which exist in less than 5% of individuals (Kremer et al. 1991; Sutherland et al. 1998). To date, over 200 CFSs (Mrasek et al. 2010) and 33 RFS (Sutherland 2003) have been identified in human cells.
CFSs are hard-to-replicate and are hotspots for genomic alterations found in human cancers. The molecular pathway underlying CFS-associated genomic instability and the reason why CFSs are hypersensitive to RS are still not fully understood. Several features of CFSs may underline their instability. First, some CFSs are naturally late replicating. FRA3B was the first CFS shown to be late replicating during S phase using fluorescence in situ hybridization (FISH) (Le Beau et al. 1998). Through labeling late-replicating DNA with BrdU in human cells, Wang et al. showed that that RS-induced breaks or gaps preferentially occurred on the allele of FRA3B that replicated later. Similar effects were observed at other CFSs studied, including FRA16D, FRA7H, FRA7G, FRA1H, and FRA2G (Palakodeti et al. 2004; Pelliccia et al. 2008). These studies provided direct evidence that allele-specific late replication is involved in the fragility of many of the most prominent CFS in human cells (Wang et al. 1999). This replication-timing analysis indicated that these CFSs either start to replicate very late or the progression of the replication machinery is unusually slow or interrupted in S phase.
CFSs are also considered to be associated with replication origin-poor loci. A study on FRA3B in lymphoblastoid cells showed that replication fork speed within FRA3B is similar to that in the bulk genome, and instead, it is the lack of replication initiation events that contribute to its fragility (Letessier et al. 2011). This might explain to some extent the known tissue specificity of CFS expression. It is known that the replication program is reset during differentiation, which leads to a defined, tissue-specific, replication timing profile (Dazy et al. 2006; Hansen et al. 2010; Hiratani et al. 2008). Indeed, FRA3B, which has a paucity of replication initiation events in lymphoblastoid cells, and is known to be highly fragile under RS in this cell type, but not in fibroblasts. Similar results have been observed with FRA16D and FRA6E (Letessier et al. 2011; Palumbo et al. 2010).
Another feature of CFSs is that DNA secondary structures can form within these regions. Early studies demonstrated that some CFSs (FRA2G, FRA3B, FRAXB, FRA7E, FRA7H, FRA8C, and FRA16D) are enriched with A/T rich sequences (e.g., AT microsatellites) that tend to adopt an abnormal DNA structure with a very high degree of flexibility and instability (Arlt et al. 2002; Boldog et al. 1997; Mishmar et al. 1998; Ohta et al. 1996). It is plausible that secondary structures located within CFSs could then perturb DNA replication fork progression. In support of this, transfer of a short AT-rich region (Flex1) derived from FRA16D into the yeast genome can cause replication stalling and DNA breakage at this region (Zhang and Freudenreich 2007). Also, in mammalian cells, CFS-derived AT-rich DNA is a hotspot for mitotic recombination (Wang et al. 2014). In addition, proteins known to counteract DNA secondary structures, such as DNA helicases (e.g., BLM) and structure-specific endonucleases (e.g., MUS81), are reported to be crucial for CFS stability, which supports the notion that DNA secondary structures might play a crucial role in the fragility of at least some CFSs (Lu et al. 2013; Pirzio et al. 2008; Shah et al. 2010; Ying et al. 2013).
Despite the above discussion, it is hard to envisage how any particular sequence feature could explain the tissue-specific nature of CFS fragility. By comparing CFSs between two different cell types, < 20% of the sites of CFS fragility are shared (Le Tallec et al. 2013). Hence, the propensity of a given CFS to be fragile varies greatly between two different cell types. For example, FRA16D is among the most fragile site in human lymphocytes, but shows much lower levels of fragility in fibroblasts (Helmrich et al. 2006; Hosseini et al. 2013; Le Tallec et al. 2011, 2013).
One interesting feature of CFSs is that they frequently co-localize with large genes, with some as large as 1–2 mega base pairs (Mb) (Smith et al. 2007). For example, the two most frequently studied CFSs, FRA16D and FRA3B, are associated with the very long WWOX (1.1 Mb) and FHIT (1.5 Mb) genes, respectively (Huang et al. 2015; Ohta et al. 1996). Indeed, a large scale CFS mapping study in epithelial and erythroid cells indicated that more than 80% of human CFSs, and 100% of CFSs in mouse embryonic fibroblasts, harbor genes over 300 kb in size, more than five times the median gene size (Le Tallec et al. 2013). It has also been reported that actively transcribed large genes are highly associated with CFS expression, as well as being hotspots for copy-number variation (CNV) (Wilson et al. 2015). As noted above, the collisions between transcription and replication that would be expected to occur more frequently in long genes can induce R-loop formation that is deleterious to replication fork progression, increasing the probability of the CFS being fragile. Consistent with this being a factor, overexpression of RNase H1, which removes R-loops, can reduce CFS expression (Helmrich et al. 2011). Moreover, the CFS marker, FANCD2, which was shown to bind to the center of large actively transcribed genes under RS (Pentzold et al. 2018), is required for resolving R-loops formed within CFSs (García-Rubio et al. 2015; Madireddy et al. 2016; Schwab et al. 2015). Because transcription is constantly reprogrammed during the cell development, and some tissue-specific transcription components are known to regulate the change of gene expression during development (Hiller et al. 2001), transcription of large genes harboring CFSs could readily explain the tissue-specific nature of CFS expression.
Rare fragile sites (RFSs) exist in less than 5% of the individuals. RFSs often contain di- or tri-nucleotide repeat sequences that are unstable and prone to expand and contract. They are sub-divided into folate-sensitive and non-folate-sensitive groups. Currently, there are 24 known folate-sensitive RFSs, all of which are associated with the expansion of a CGG-repeat sequence (Durkin and Glover 2007). To date, FRAXA is the best studied folate-sensitive RFS in humans because of its association with defined pathological disorders (Kidd et al. 2014). The expansion of the CGG-repeat element within the 5′ untranslated region (5′UTR) of the human FMR1 gene at FRAXA is responsible for disorders that arise through different mechanisms: FMR1 gene silencing gives rise to fragile X syndrome (requiring > 200 CGG repeats), whereas toxic RNA stabilization induces an ovarian insufficiency disorder or a neurological disorder associated with ataxia where there are between 50 and 200 CGG repeats (Bell et al. 1991; Hansen et al. 1992; Santoro et al. 2012; Sherman 2000). Using DNA:RNA immunoprecipitation (DRIP) of genomic DNA from cells with either normal (∼ 30 CGGs) or pre-mutation FRAXA alleles (55-200 CGGs), Loomis et al. reported that transcription through the GC-rich FMR1 5′UTR region favors R-loop formation. It is plausible that, in those cells with either normal or pre-mutation FRAXA alleles, the formation of R-loops (and possibly other DNA secondary structures) at the transcribed FMR1 gene could generate an obstacle for the DNA replication machinery. This might then lead to the activation of a group of DNA-damage proteins, which could either promote replication re-start or inadvertently trigger the abnormal expansion of the CGG repeats during attempted repair (Loomis et al. 2014). Further research is clearly warranted in this direction. Interestingly, by analyzing a cell line with a mutant FRAXA allele (> 500 CGGs), Groh et al. demonstrated that R-loops act as an initial trigger to promote FMR1 gene silencing, possibly by their association with the repressive H3K9me2 chromatin mark (Groh et al. 2014).
There is also a group of fragile sites that are designated as being ‘early replicating’ (called ‘early replicating fragile sites’; ERFSs) (Barlow et al. 2013). These are fragile sites that tend to overlap with highly transcribed, early replicating gene clusters. In addition, ERFSs often lie close to replication origins, which might increase the probability of replication–transcription collisions (Barlow et al. 2013; Mortusewicz et al. 2013). Importantly, like CFSs, the fragility of ERFSs can also be stimulated by oncogene overexpression. It is thought that transcription occurring in the G1 phase serves to inactivate replication origins within the body of many transcribed genes, thus reducing the potential for conflicts (Lemmens et al. 2018). Foreshortening of G1 caused by oncogene overexpression, therefore, leaves insufficient time for transcription of genes, and, subsequently, increases the chances of replication–transcription conflicts (Halazonetis et al. 2008; Macheret and Halazonetis 2018).
Highly transcribed regions
In human cells, rRNA and tRNA genes are highly transcribed by RNAPI or RNAPIII, respectively, to meet the demand for protein synthesis. Therefore, the chances of the replication and transcription machineries colliding with each other should be particularly high at these loci (Deshpande and Newlon 1996; Kobayashi 2014; Takeuchi et al. 2003). As mentioned earlier, however, replication fork barriers are a feature of rDNA and tDNA loci to limit head-on collisions. Nonetheless, collisions associated with R loops are seen commonly at rDNA/tDNA genes (Chen et al. 2017; Lindström et al. 2018). Any deleterious consequences of such collisions are seemingly limited by the action of RNase H1, PIF1, and TOP1 (El Hage et al. 2010, 2014; Shen et al. 2017; Tran et al. 2017).
Telomeres
Telomeres are the repetitive DNA sequences located at the ends of chromosomes. Telomeres also exhibit fragility, as shown by the discontinuity or loss of signals when probing for telomeric DNA using FISH (Özer et al. 2018). Telomere fragility is also exacerbated by a low dose of APH, in the manner used for induction of CFS fragility, supporting the notion that telomeres are hard-to-replicate and should be viewed as a class of specialized fragile sites (Sfeir et al. 2009). Although telomeres are largely heterochromatic, transcription can be initiated in the sub-telomeric region to generate a transcript called TERRA (telomeric repeat-containing RNA). TERRA can form stable R-loops with telomeric DNA (Azzalin et al. 2007), and indeed, these have been observed in many eukaryotic cell types (Rippe and Luke 2015). As discussed above, R-loops are challenging obstacles for replication forks and among the major sources of RS. On top of that, the displaced, G-rich single-stranded DNA formed during TERRA R-loop formation favors the stabilization of G-quadruplexes. Thus, TERRA R loops may be a major source of RS at telomeres. The key enzyme in resolving R-loops, RNase H1, is known to regulate telomere length in telomerase-negative ALT cells that express elevated levels of TERRA (Arora et al. 2014). The FEN1 flap endonuclease is also known to process DNA–RNA hybrids to avoid telomere fragility (Teasley et al. 2015), while loss of the UPF1 helicase leads to TERRA accumulation and impaired telomeric leading stand DNA synthesis (Azzalin et al. 2007). Intriguingly, in human telomerase-positive cells and in yeast cells, TERRA abundance is cell-cycle-dependent, with the lowest level in the S and G2 phases (Flynn et al. 2015). This is probably one of the strategies cells have evolved to avoid a collision with the replication fork within telomeres.
Concluding remarks
DNA replication and RNA transcription are two processes that are essential for cell proliferation. Ironically, because these two processes are competing for the same template DNA, conflicts between them are inevitable. From bacteria to mammals, all cells have adopted strategies to minimize these conflicts or resolve them faithfully if they happen to occur. Recent lines of evidence indicate that replication–transcription conflicts represent perhaps the major source of intrinsic genomic instability, which has the potential to drive human disease. Therefore, it is crucial to decipher the processes underlying the cause of these conflicts, and the mechanisms used to ameliorate conflicts when they occur.
In the past two decades, one of the most significant advances in this field is the recognition of the importance of R-loops in driving RS and genome instability (Drolet et al. 1995). If these structures are not resolved in a timely manner, they can form obstacles for DNA replication, and present serious problems for genome maintenance. These advances in R-loop biology have been greatly facilitated by the development of an antibody (S9.6) that can bind to R-loop structures (Boguslawski et al. 1986). This permits detecting R-loops directly in cells using immunofluorescence (Sollier et al. 2014), although there are caveats about its specificity in this context, and the identification of genomic regions that form R-loops using DRIP coupled with high-throughput sequencing (Halász et al. 2017). Such analysis has also permitted the identification not only of the epigenetic changes in the vicinity of R-loops (Chen et al. 2015; Ginno et al. 2013; Sanz et al. 2016), but also, when coupled to mass spectrometry, of the proteins bound to R-loops (Cristini et al. 2018). These technological advances will also greatly facilitate research in the coming years to address the following questions (amongst many others): (1) How do R-loops contribute positively to physiological processes such as regulation of gene expression and the initiation of DSB repair (Ohle et al. 2016)? Do these putative functions occur mainly through the recruitment of R-loop-binding proteins? (2) In cancer cells where there is an elevated frequency of transcription–replication conflicts at specific hotspots like CFSs, which helicases/nucleases are employed to promote R-loop resolution? (3) When R-loops cannot be resolved, how is a DNA-damage response initiated, and what are the downstream consequences of such a response? Given the interest in the basic biology of DNA replication and transcription, and the emerging role played by replication–transcription in human disease, we do not anticipate any diminution in activity in this field in the coming years.
References
Aguilera, A., & García-Muse, T. (2012). R Loops: From transcription byproducts to threats to genome stability. Molecular Cell,46, 115–124.
Almeida, R., Fernández-Justel, J. M., Santa-María, C., Cadoret, J.-C., Cano-Aroca, L., Lombraña, R., et al. (2018). Chromatin conformation regulates the coordination between DNA replication and transcription. Nature Communications,9, 1590.
Arlt, M. F., Miller, D. E., Beer, D. G., & Glover, T. W. (2002). Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes, Chromosomes & Cancer,33, 82–92.
Arora, R., Lee, Y., Wischnewski, H., Brun, C. M., Schwarz, T., & Azzalin, C. M. (2014). RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature Communications,5, 5220.
Artsimovitch, I., & Landick, R. (2000). Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proceedings of the National Academy of Sciences of the United States of America,97, 7090–7095.
Azvolinsky, A., Giresi, P. G., Lieb, J. D., & Zakian, V. A. (2009). Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Molecular Cell,34, 722–734.
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., & Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science (New York, NY),318, 798–801.
Barlow, J. H., Faryabi, R. B., Callen, E., Wong, N., Malhowski, A., Chen, H. T., et al. (2013). Identification of early replicating fragile sites that contribute to genome instability. Cell,152, 620–632.
Bell, M., Hirst, M., Nakahori, Y., MacKinnon, R., Roche, A., Flint, T., et al. (1991). Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell,64, 861–866.
Belotserkovskii, B. P., & Hanawalt, P. C. (2011). Anchoring nascent RNA to the DNA template could interfere with transcription. Biophysical Journal,100, 675–684.
Belotserkovskii, B. P., Liu, R., Tornaletti, S., Krasilnikova, M. M., Mirkin, S. M., & Hanawalt, P. C. (2010). Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proceedings of the National Academy of Sciences of the United States of America,107, 12816–12821.
Belotserkovskii, B. P., Soo Shin, J. H., & Hanawalt, P. C. (2017). Strong transcription blockage mediated by R-loop formation within a G-rich homopurine–homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Research,45, 6589–6599.
Bentley, D. L. (2014). Coupling mRNA processing with transcription in time and space. Nature Reviews Genetics,15, 163–175.
Bermejo, R., Doksani, Y., Capra, T., Katou, Y.-M., Tanaka, H., Shirahige, K., et al. (2007). Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes & Development,21, 1921–1936.
Bermejo, R., Lai, Mong S., & Foiani, M. (2012). Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Molecular Cell,45, 710–718.
Bertoli, C., Herlihy, A. E., Pennycook, B. R., Kriston-Vizi, J., & de Bruin, R. A. M. (2016). Sustained E2F-dependent transcription is a key mechanism to prevent replication-stress-induced DNA DAMAGE. Cell Rep,15, 1412–1422.
Bertoli, C., Skotheim, J. M., & de Bruin, R. A. M. (2013). Control of cell cycle transcription during G1 and S phases. Nature Reviews Molecular Cell Biology,14, 518–528.
Bleichert, F., Botchan, M. R., & Berger, J. M. (2017). Mechanisms for initiating cellular DNA replication. Science,. https://doi.org/10.1126/science.aah6317.
Boguslawski, S. J., Smith, D. E., Michalak, M. A., Mickelson, K. E., Yehle, C. O., Patterson, W. L., et al. (1986). Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods,89, 123–130.
Boldog, F., Gemmill, R. M., West, J., Robinson, M., Robinson, L., Li, E., et al. (1997). Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet,6, 193–203.
Boos, D., Yekezare, M., & Diffley, J. F. (2013). Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science (New York, NY),340, 981–984.
Cadoret, J. C., Meisch, F., Hassan-Zadeh, V., Luyten, I., Guillet, C., Duret, L., et al. (2008). Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proceedings of the National Academy of Sciences of the United States of America,105, 15837–15842.
Candelli, T., Gros, J., & Libri, D. (2018). Pervasive transcription fine-tunes replication origin activity. eLife,7, e40802.
Castel, Stephane E., Ren, J., Bhattacharjee, S., Chang, A.-Y., Sánchez, M., Valbuena, A., et al. (2014). Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell,159, 572–583.
Chen, Y.-Z., Bennett, C. L., Huynh, H. M., Blair, I. P., Puls, I., Irobi, J., et al. (2004). DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet,74, 1128–1135.
Chen, P. B., Chen, H. V., Acharya, D., Rando, O. J., & Fazzio, T. G. (2015). R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol,22, 999–1007.
Chen, L., Chen, J.-Y., Zhang, X., Gu, Y., Xiao, R., Shao, C., et al. (2017). R-chip using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Molecular Cell,68, 745–757.e745.
Chen, Y.-H., Keegan, S., Kahli, M., Tonzi, P., Fenyö, D., Huang, T. T., et al. (2019). Transcription shapes DNA replication initiation and termination in human cells. Nat Struct Mol Biol,26, 67–77.
Cheung, A. C. M., & Cramer, P. (2011). Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature,471, 249–253.
Churchman, L. S., & Weissman, J. S. (2011). Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature,469, 368–373.
Cornacchia, D., Dileep, V., Quivy, J.-P., Foti, R., Tili, F., Santarella-Mellwig, R., et al. (2012). Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J,31, 3678–3690.
Cristini, A., Groh, M., Kristiansen, M. S., & Gromak, N. (2018). RNA/DNA hybrid interactome identifies dxh9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Reports,23, 1891–1905.
Dazy, S., Gandrillon, O., Hyrien, O., & Prioleau, M. N. (2006). Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Reports,7, 806–811.
Deshpande, A. M., & Newlon, C. S. (1996). DNA replication fork pause sites dependent on transcription. Science (New York, NY),272, 1030–1033.
Dimitrova, D. S. (2011). DNA replication initiation patterns and spatial dynamics of the human ribosomal RNA gene loci. Journal of Cell Science,124, 2743.
Dimova, D. K., & Dyson, N. J. (2005). The E2F transcriptional network: old acquaintances with new faces. Oncogene,24, 2810–2826.
Drolet, M., Phoenix, P., Menzel, R., Massé, E., Liu, L. F., & Crouch, R. J. (1995). Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: r-loop formation is a major problem in the absence of DNA topoisomerase I. Proceedings of the National Academy of Sciences,92, 3526–3530.
Durkin, S. G., & Glover, T. W. (2007). Chromosome fragile sites. Annual Review of Genetics,41, 169–192.
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E., & Nudler, E. (2011). Linking RNA polymerase backtracking to genome instability in E. coli. Cell,146, 533–543.
El Hage, A., French, S. L., Beyer, A. L., & Tollervey, D. (2010). Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & Development,24, 1546–1558.
El Hage, A., Webb, S., Kerr, A., & Tollervey, D. (2014). Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genetics,10, e1004716.
Epshtein, V., Kamarthapu, V., McGary, K., Svetlov, V., Ueberheide, B., Proshkin, S., et al. (2014). UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature,505, 372–377.
Flynn, R. L., Cox, K. E., Jeitany, M., Wakimoto, H., Bryll, A. R., Ganem, N. J., et al. (2015). Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science (New York, NY),347, 273–277.
Foti, R., Gnan, S., Cornacchia, D., Dileep, V., Bulut-Karslioglu, A., Diehl, S., et al. (2016). Nuclear architecture organized by Rif1 underpins the replication-timing program. Molecular Cell,61, 260–273.
Ganesan, A., Spivak, G., & Hanawalt, P. C. (2012). Chapter 2—transcription-coupled DNA repair in prokaryotes. In P. W. Doetsch (Ed.), Progress in molecular biology and translational science (pp. 25–40). Cambridge, MA, US: Academic Press.
Garcia-Muse, T., & Aguilera, A. (2016). Transcription-replication conflicts: how they occur and how they are resolved. Nature Reviews Molecular Cell Biology,17, 553–563.
García-Rubio, M. L., Pérez-Calero, C., Barroso, S. I., Tumini, E., Herrera-Moyano, E., Rosado, I. V., et al. (2015). The fanconi anemia pathway protects genome integrity from R-loops. PLOS Genetics,11, e1005674.
Gerber, J. K., Gogel, E., Berger, C., Wallisch, M., Muller, F., Grummt, I., et al. (1997). Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell,90, 559–567.
Ginno, P. A., Lim, Y. W., Lott, P. L., Korf, I., & Chedin, F. (2013). GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Research,23, 1590–1600.
Gomez-Herreros, F., de Miguel-Jimenez, L., Millan-Zambrano, G., Penate, X., Delgado-Ramos, L., Munoz-Centeno, M. C., et al. (2012). One step back before moving forward: regulation of transcription elongation by arrest and backtracking. FEBS Letters,586, 2820–2825.
Groh, M., Lufino, M. M., Wade-Martins, R., & Gromak, N. (2014). R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genetics,10, e1004318.
Guy, L., & Roten, C.-A. H. (2004). Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene,340, 45–52.
Halász, L., Karányi, Z., Boros-Oláh, B., Kuik-Rózsa, T., Sipos, É., Nagy, É., et al. (2017). RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Research,27, 1063–1073.
Halazonetis, T. D., Gorgoulis, V. G., & Bartek, J. (2008). An oncogene-induced DNA damage model for cancer development. Science (New York, NY),319, 1352–1355.
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T., & Cimprich, K. A. (2017). Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell,170, 774–786.e719.
Hansen, R. S., Gartler, S. M., Scott, C. R., Chen, S.-H., & xLaird, C. M. (1992). Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Human Molecular Genetics,1, 571–578.
Hansen, R. S., Thomas, S., Sandstrom, R., Canfield, T. K., Thurman, R. E., Weaver, M., et al. (2010). Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proceedings of the National Academy of Sciences of the United States of America,107, 139–144.
Hatchi, E., Skourti-Stathaki, K., Ventz, S., Pinello, L., Yen, A., Kamieniarz-Gdula, K., et al. (2015). BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Molecular Cell,57, 636–647.
Helmrich, A., Ballarino, M., Nudler, E., & Tora, L. (2013). Transcription-replication encounters, consequences and genomic instability. Nature Structural and Molecular Biology,20, 412–418.
Helmrich, A., Ballarino, M., & Tora, L. (2011). Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Molecular Cell,44, 966–977.
Helmrich, A., Stout-Weider, K., Hermann, K., Schrock, E., & Heiden, T. (2006). Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Research,16, 1222–1230.
Herlihy, A. E., & de Bruin, R. A. M. (2017). The role of the transcriptional response to DNA replication stress. Genes (Basel),8, 92.
Herrgård, M. J., Lee, B.-S., Portnoy, V., & Palsson, B. Ø. (2006). Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Research,16, 627–635.
Hiller, M. A., Lin, T. Y., Wood, C., & Fuller, M. T. (2001). Developmental regulation of transcription by a tissue-specific TAF homolog. Genes & Development,15, 1021–1030.
Hiratani, I., Ryba, T., Itoh, M., Yokochi, T., Schwaiger, M., Chang, C. W., et al. (2008). Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biology,6, e245.
Hosseini, S. A., Horton, S., Saldivar, J. C., Miuma, S., Stampfer, M. R., Heerema, N. A., et al. (2013). Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes, Chromosomes and Cancer,52, 1017–1029.
Huang, Y. H., Chang, N. S., & Tseng, S. H. (2015). Expression of WW domain-containing oxidoreductase WWOX in pterygium. Molecular Vision,21, 711–717.
Huertas, P., & Aguilera, A. (2003). Cotranscriptionally formed DNA: RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular Cell,12, 711–721.
Ivessa, A. S., Lenzmeier, B. A., Bessler, J. B., Goudsouzian, L. K., Schnakenberg, S. L., & Zakian, V. A. (2003). The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Molecular Cell,12, 1525–1536.
Kidd, S. A., Lachiewicz, A., Barbouth, D., Blitz, R. K., Delahunty, C., McBrien, D., et al. (2014). Fragile X syndrome: a review of associated medical problems. Pediatrics,134, 995–1005.
Knott, Simon R. V., Peace, Jared M., Ostrow, A. Z., Gan, Y., Rex, Alexandra E., Viggiani, Christopher J., et al. (2012). Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell,148, 99–111.
Kobayashi, T. (2014). Ribosomal RNA gene repeats, their stability and cellular senescence. Proceedings of the Japan Academy, Series B,90, 119–129.
Kremer, E. J., Pritchard, M., Lynch, M., Yu, S., Holman, K., Baker, E., et al. (1991). Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science (New York, NY),252, 1711–1714.
Le Beau, M. M., Rassool, F. V., Neilly, M. E., Espinosa, R., 3rd, Glover, T. W., Smith, D. I., et al. (1998). Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Human Molecular Genetics,7, 755–761.
Le Tallec, B., Dutrillaux, B., Lachages, A.-M., Millot, G. A., Brison, O., & Debatisse, M. (2011). Molecular profiling of common fragile sites in human fibroblasts. Nature Structural and Molecular Biology,18, 1421–1423.
Le Tallec, B., Millot, G. A., Blin, M. E., Brison, O., Dutrillaux, B., & Debatisse, M. (2013). Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Reports,4, 420–428.
Lemmens, B., Hegarat, N., Akopyan, K., Sala-Gaston, J., Bartek, J., Hochegger, H., et al. (2018). DNA replication determines timing of mitosis by restricting CDK1 and PLK1 activation. Molecular Cell,71, 117–128.
Letessier, A., Millot, G. A., Koundrioukoff, S., Lachagès, A.-M., Vogt, N., Hansen, R. S., et al. (2011). Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature,470, 120–123.
Li, T. K., Chen, A. Y., Yu, C., Mao, Y., Wang, H., & Liu, L. F. (1999). Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes & Development,13, 1553–1560.
Li, X., & Manley, J. L. (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell,122, 365–378.
Lin, Y.-L., & Pasero, P. (2012). Interference between DNA replication and transcription as a cause of genomic instability. Current Genomics,13, 65–73.
Lindström, M. S., Jurada, D., Bursac, S., Orsolic, I., Bartek, J., & Volarevic, S. (2018). Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene,37, 2351–2366.
Little, R. D., Platt, T. H., & Schildkraut, C. L. (1993). Initiation and termination of DNA replication in human rRNA genes. Molecular and Cell Biology,13, 6600–6613.
Liu, B., Lie Wong, M., Tinker, R. L., Geiduschek, E. P., & Alberts, B. M. (1993). The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature,366, 33–39.
Lockhart, A., Pires, V. B., Bento, F., Kellner, V., Luke-Glaser, S., Yakoub, G., et al. (2019). RNase H1 and H2 are differentially regulated to process RNA-DNA hybrids. Cell Reports,29, 2890–2900.e2895.
Loomis, E. W., Sanz, L. A., Chedin, F., & Hagerman, P. J. (2014). Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genetics,10, e1004294.
Lu, X., Parvathaneni, S., Hara, T., Lal, A., & Sharma, S. (2013). Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Molecular Cancer,12, 29.
Macheret, M., & Halazonetis, T. D. (2018). Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature,555, 112–116.
Madireddy, A., Kosiyatrakul, S. T., Boisvert, R. A., Herrera-Moyano, E., Garcia-Rubio, M. L., Gerhardt, J., et al. (2016). FANCD2 facilitates replication through common fragile sites. Molecular Cell,64, 388–404.
Marchal, C., Sima, J., & Gilbert, D. M. (2019). Control of DNA replication timing in the 3D genome. Nature Reviews Molecular Cell Biology,20, 721–737.
Merrikh, H., Zhang, Y., Grossman, A. D., & Wang, J. D. (2012). Replication–transcription conflicts in bacteria. Nature Reviews Microbiology,10, 449–458.
Meryet-Figuiere, M., Alaei-Mahabadi, B., Ali, M. M., Mitra, S., Subhash, S., Pandey, G. K., et al. (2014). Temporal separation of replication and transcription during S-phase progression. Cell Cycle (Georgetown, Tex),13, 3241–3248.
Miotto, B., Ji, Z., & Struhl, K. (2016). Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proceedings of the National Academy of Sciences,113, E4810–E4819.
Mishmar, D., Rahat, A., Scherer, S. W., Nyakatura, G., Hinzmann, B., Kohwi, Y., et al. (1998). Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proceedings of the National Academy of Sciences,95, 8141–8146.
Moreira, M.-C., Klur, S., Watanabe, M., Németh, A. H., Ber, I. L., Moniz, J.-C., et al. (2004). Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nature Genetics,36, 225–227.
Mortusewicz, O., Herr, P., & Helleday, T. (2013). Early replication fragile sites: where replication-transcription collisions cause genetic instability. The EMBO Journal,32, 493–495.
Mrasek, K., Schoder, C., Teichmann, A.-C., Behr, K., Franze, B., Wilhelm, K., et al. (2010). Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. International Journal of Oncology,36, 929–940.
Muller, M., Lucchini, R., & Sogo, J. M. (2000). Replication of yeast rDNA initiates downstream of transcriptionally active genes. Molecular Cell,5, 767–777.
Murakami, H., Aiba, H., Nakanishi, M., & Murakami-Tonami, Y. (2010). Regulation of yeast forkhead transcription factors and FoxM1 by cyclin-dependent and polo-like kinases. Cell Cycle (Georgetown, Tex),9, 3253–3262.
Nudler, E. (2012). RNA polymerase backtracking in gene regulation and genome instability. Cell,149, 1438–1445.
Ohle, C., Tesorero, R., Schermann, G., Dobrev, N., Sinning, I., & Fischer, T. (2016). Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell,167, 1001–1013.e1007.
Ohta, M., Inoue, H., Cotticelli, M. G., Kastury, K., Baffa, R., Palazzo, J., et al. (1996). The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell,84, 587–597.
Opalka, N., Chlenov, M., Chacon, P., Rice, W. J., Wriggers, W., & Darst, S. A. (2003). Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell,114, 335–345.
Özer, Ö., Bhowmick, R., Liu, Y., & Hickson, I. D. (2018). Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget,9, 15836–15846.
Palakodeti, A., Han, Y., Jiang, Y., & Le Beau, M. M. (2004). The role of late/slow replication of the FRA16D in common fragile site induction. Genes, Chromosomes & Cancer,39, 71–76.
Palumbo, E., Matricardi, L., Tosoni, E., Bensimon, A., & Russo, A. (2010). Replication dynamics at common fragile site FRA6E. Chromosoma,119, 575–587.
Pasero, P., Bensimon, A., & Schwob, E. (2002). Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Development,16, 2479–2484.
Pelliccia, F., Bosco, N., Curatolo, A., & Rocchi, A. (2008). Replication timing of two human common fragile sites: fRA1H and FRA2G. Cytogenet Genome Research,121, 196–200.
Pentzold, C., Shah, S. A., Hansen, N. R., Le Tallec, B., Seguin-Orlando, A., Debatisse, M., et al. (2018). FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells. Nucl Acids Res,46, 1280–1294.
Pirzio, L. M., Pichierri, P., Bignami, M., & Franchitto, A. (2008). Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol,180, 305–314.
Pomerantz, R. T., & O’Donnell, M. (2008). The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature,456, 762–766.
Pope, B. D., Ryba, T., Dileep, V., Yue, F., Wu, W., Denas, O., et al. (2014). Topologically associating domains are stable units of replication-timing regulation. Nature,515, 402–405.
Prado, F., & Aguilera, A. (2005). Impairment of replication fork progression mediates RNA polII transcription-associated recombination. THE EMBO Journal,24, 1267–1276.
Proshkin, S., Rahmouni, A. R., Mironov, A., & Nudler, E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science (New York, NY),328, 504–508.
Richard, P., Feng, S., & Manley, J. L. (2013). A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes & Development,27, 2227–2232.
Rippe, K., & Luke, B. (2015). TERRA and the state of the telomere. Nat Struct Mol Biol,22, 853–858.
Rocha, E. P., & Danchin, A. (2003). Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nature Genetics,34, 377–378.
Santoro, M. R., Bray, S. M., & Warren, S. T. (2012). Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annual Review of Pathology: Mechanisms of Disease,7, 219–245.
Santos-Pereira, J. M., & Aguilera, A. (2015). R loops: new modulators of genome dynamics and function. Nature Reviews Genetics,16, 583–597.
Sanz, L. A., Hartono, S. R., Lim, Y. W., Steyaert, S., Rajpurkar, A., Ginno, P. A., et al. (2016). Prevalent, dynamic, and conserved r-loop structures associate with specific epigenomic signatures in mammals. Molecular Cell,63, 167–178.
Saponaro, M., Kantidakis, T., Mitter, R., Kelly, Gavin P., Heron, M., Williams, H., et al. (2014). RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell,157, 1037–1049.
Schwab, R. A., Nieminuszczy, J., Shah, F., Langton, J., Lopez Martinez, D., Liang, C.-C., et al. (2015). The fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Molecular Cell,60, 351–361.
Sequeira-Mendes, J., Díaz-Uriarte, R., Apedaile, A., Huntley, D., Brockdorff, N., & Gómez, M. (2009). Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genetics,5, e1000446.
Sfeir, A., Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., et al. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell,138, 90–103.
Shah, S. N., Opresko, P. L., Meng, X., Lee, M. Y., & Eckert, K. A. (2010). DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Research,38, 1149–1162.
Shen, W., Sun, H., De Hoyos, C. L., Bailey, J. K., Liang, X. H., & Crooke, S. T. (2017). Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Researchearch,45, 10672–10692.
Sherman, S. L. (2000). Premature ovarian failure among fragile X premutation carriers: parent-of-origin effect? AmJ Hum Genet,67, 11–13.
Sima, J., Chakraborty, A., Dileep, V., Michalski, M., Klein, K. N., Holcomb, N. P., et al. (2019). Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell,176, 816–830.e818.
Smith, D. I., McAvoy, S., Zhu, Y., & Perez, D. S. (2007). Large common fragile site genes and cancer. Seminars in Cancer Biology,17, 31–41.
Sollier, J., Stork, Caroline T., García-Rubio, María L., Paulsen, Renee D., Aguilera, A., & Cimprich, Karlene A. (2014). Transcription-Coupled nucleotide excision repair factors promote R-loop-induced genome instability. Molecular Cell,56, 777–785.
Sutherland, G. R. (1979). Heritable fragile sites on human chromosomes I. Factors affecting expression in lymphocyte culture. American Journal of Human Genetics,31, 125–135.
Sutherland, G. (2003). Rare fragile sites. Cytogenet Genome Research,100, 77–84.
Sutherland, G. R., Baker, E., & Richards, R. I. (1998). Fragile sites still breaking. Trends Genet,14, 501–506.
Takeuchi, Y., Horiuchi, T., & Kobayashi, T. (2003). Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes & Development,17, 1497–1506.
Tazumi, A., Fukuura, M., Nakato, R., Kishimoto, A., Takenaka, T., Ogawa, S., et al. (2012). Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes & Development,26, 2050–2062.
Teasley, D. C., Parajuli, S., Nguyen, M., Moore, H. R., Alspach, E., Lock, Y. J., et al. (2015). Flap endonuclease 1 limits telomere fragility on the leading strand. The Journal of Biological Chemistry,290, 15133–15145.
Thomas, M. J., Platas, A. A., & Hawley, D. K. (1998). Transcriptional fidelity and proofreading by RNA polymerase II. Cell,93, 627–637.
Thu, Y. M., & Bielinsky, A. K. (2013). Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci,38, 184–194.
Tran, P. L. T., Pohl, T. J., Chen, C. F., Chan, A., Pott, S., & Zakian, V. A. (2017). PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nature Communications,8, 15025.
Tuduri, S., Crabbé, L., Conti, C., Tourrière, H., Holtgreve-Grez, H., Jauch, A., et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature Cell Biology,11, 1315–1324.
Urban, V., Dobrovolna, J., Huhn, D., Fryzelkova, J., Bartek, J., & Janscak, P. (2016). RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. The Journal of Cell Biology,214, 401–415.
Vermeulen, W., & Fousteri, M. (2013). Mammalian transcription-coupled excision repair. Cold Spring Harbor Perspectives in Biology,5, a012625–a012625.
Wang, L., Darling, J., Zhang, J.-S., Huang, H., Liu, W., & Smith, D. I. (1999). Allele-specific Late replication and fragility of the most active common fragile site, FRA3B. Human Molecular Genetics,8, 431–437.
Wang, H., Li, Y., Truong, L. N., Shi, L. Z., Hwang, P. Y., He, J., et al. (2014). CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Molecular Cell,54, 1012–1021.
Wei, X., Samarabandu, J., Devdhar, R. S., Siegel, A. J., Acharya, R., & Berezney, R. (1998). Segregation of transcription and replication sites into higher order domains. Science (New York, NY),281, 1502–1505.
Wilson, T. E., Arlt, M. F., Park, S. H., Rajendran, S., Paulsen, M., Ljungman, M., et al. (2015). Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Research,25, 189–200.
Wilson, Marcus D., Harreman, M., Taschner, M., Reid, J., Walker, J., Erdjument-Bromage, H., et al. (2013). Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell,154, 983–995.
Wittenberg, C., & Reed, S. I. (2005). Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene,24, 2746–2755.
Yamazaki, S., Hayano, M., & Masai, H. (2013). Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet,29, 449–460.
Yamazaki, S., Ishii, A., Kanoh, Y., Oda, M., Nishito, Y., & Masai, H. (2012). Rif1 regulates the replication timing domains on the human genome. THE EMBO Journal,31, 3667–3677.
Ying, S., Minocherhomji, S., Chan, K. L., Palmai-Pallag, T., Chu, W. K., Wass, T., et al. (2013). MUS81 promotes common fragile site expression. Nature Cell Biology,15, 1001–1007.
Zaratiegui, M., Castel, S. E., Irvine, D. V., Kloc, A., Ren, J., Li, F., et al. (2011). RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature,479, 135–138.
Zhang, H., & Freudenreich, C. H. (2007). An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Molecular Cell,27, 367–379.
Zhu, W., Ukomadu, C., Jha, S., Senga, T., Dhar, S. K., Wohlschlegel, J. A., et al. (2007). Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes & Development,21, 2288–2299.
Acknowledgements
This work was supported by grants from the Chinese Scholarship Council (PhD fellowship 201406170048; WW), the Nordea Foundation of Denmark (IDH), The Danish National Research Foundation (DNRF115; IDH and YL), and the European Commission (H2020/Marie Skłodowska-Curie, 859853; IDH and YL).
Author information
Authors and Affiliations
Contributions
WW, IDH, and YL wrote and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, W., Hickson, I.D. & Liu, Y. The prevention and resolution of DNA replication–transcription conflicts in eukaryotic cells. GENOME INSTAB. DIS. 1, 114–128 (2020). https://doi.org/10.1007/s42764-020-00012-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42764-020-00012-z