Abstract
Purpose
A new method of using a convolutional neural network (CNN) to perform automatic tumor segmentation from two-dimensional transaxial slices of positron emission tomography (PET) images of high-risk primary prostate cancer patients is introduced.
Methods
We compare three different methods including (1) usual image segmentation with a CNN whose continuous output is converted to binary labels with a constant threshold, (2) our new technique of choosing separate thresholds for each image PET slice with a CNN to label the pixels directly from the PET slices, and (3) the combination of the two former methods based on using the second CNN to choose the optimal thresholds to convert the output of the first CNN. The CNNs are trained and tested multiple times by using a data set of 864 slices from the PET images of 78 prostate cancer patients.
Results
According to our results, the Dice scores computed from the predictions of the second method are statistically higher than those of the typical image segmentation (p-value<0.002).
Conclusion
The new method of choosing unique thresholds to convert the pixels of the PET slices directly into binary tumor masks is not only faster and more computationally efficient but also yields better results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer is one of the most common cancer diseases, with circa 1.4 million new cases estimated worldwide yearly (Global Cancer Observatory 2022). The cancer typically progresses very slowly causing either no symptoms at all or minor problems with urination, but it can develop into a metastatic cancer by spreading into the bones of the spine and pelvis (Rawla 2019). The treatment options include surgery, radiotherapy, hormonal therapy, and active surveillance (Sciarra et al. 2018).
To image the prostate patients, a nuclear medicine imaging method known as positron emission tomography (PET) is commonly used. During the imaging, a patient is first injected with radioactive tracer substance, the positrons emitting from the unstable tracer collide with the own electrons of the human body, and a PET scanner detects the gamma quanta produced in the electron-positron annihilation. The resulting PET image shows how the radioactive substance has spread into the patient’s body, and by choosing such tracer substance accumulates in the cancerous cells due to their higher activity, cancer tumors can be detected (Townsend 2004).
One routine task required for interpreting PET images of cancer patients is tumor segmentation, which means partitioning image into two or more segments so that areas depicting cancer tissue can be labeled as positive and the rest as negative. It is often required that a medical expert divides each three-dimensional PET image into transaxial slices and considers these slices separately. Since this is time-consuming and repetitive work, different learning techniques have been developed to assist in it Blanc-Durand et al. (2018); Guo et al. (2018); Kostyszyn et al. (2021); Liedes et al. (2023); Sharif et al. (2010). Especially, a convolutional neural network (CNN) is well-suited for processing medical images because, when receiving image data, the CNN can understand the spatial relationships of the input. When one gives a PET image to a CNN designed for image segmentation, the CNN returns a matrix of the same size, and by choosing a suitable threshold, the numbers in the output can be converted into binary digits that correspond with the labels of each pixel.
However, comparing the predicted binary masks and the PET images raises the question whether it is possible to create the mask directly from the original PET image by choosing a suitable threshold to classify the pixels. Namely, cancerous tissue contains more tracer substance, and its pixels therefore have higher values in the PET image than their background, and if the image is restricted to the prostate only, there are no other regions with high concentration of tracer that would produce false positive values. In some cases, it is possible that the tracer concentration in full bladder hides the tumoral activity, but the patients are typically instructed to empty their bladder before imaging. To work properly, this method would require finding individual thresholds for each transaxial slice, but this task can be also performed with a CNN. Because it is possible that there are background negative pixels with higher values than pixels with cancer, even the best thresholds would not always result in perfect accuracy, but this unavoidable error is minimal compared to the inaccuracies of a CNN performing regular segmentation. Furthermore, this error could be fully avoided by giving the CNN finding thresholds the output of the CNN performing segmentation.
In this article, we compare three methods using a CNN to perform tumor segmentation for transaxial slices from the PET images of prostate cancer patients, including (1) typical image segmentation with a CNN whose continuous output is converted to binary labels with a constant threshold, (2) choosing a unique threshold for each image PET slice with a CNN in order to label the pixels directly from these PET slices, and (3) combining the two former procedures by giving the continuous output of the first CNN to the second CNN that chooses the optimal thresholds to convert it into binary.
Materials and methods
Software requirements
The CNNs were built and evaluated by using Python (version: 3.9.9) (van Rossum and Drake 2009) with packages TensorFlow (version: 2.7.0) (Abadi et al. 2015), Keras (version: 2.7.0) (Chollet et al. 2015), and SciPy (Virtanen et al. 2020) (version: 1.7.3), and the data was visually inspected with a medical imaging processing tool called Carimas (version: 2.10) (Rainio et al. 2023).
Data
The data of this study was collected from 78 male patients with prostate cancer, who visited Turku PET Centre in Turku, Finland, for imaging during the time from March 2018 to June 2019. The mean age of the patients was 70.9 years with a standard deviation of 7.59 years, and all the patients were treatment naive. Some of these patients had bone and lymph node metastases, but here, we focus only on the cancer tissue of the primary tumor in the prostate. All participants were at least 18 years of age and consented to research use of their data, and this study was approved by the Ethics Committee of the Hospital District of Southwest Finland (research permission number: 11065). The same data is used in the articles (Anttinen et al. 2021; Malaspina et al. 2021), which offer more medical details about the patients.
Each patient was first given a dosage of \(^{18}\)F-prostate-specific membrane antigen-1007 (\(^{18}\)F-PSMA-1007), after which a combined PET and computed tomography (CT) scan was performed with Discovery MI digital PET/CT system (GE Healthcare, Milwaukee, WI, USA). Due to the size limitations of this camera, the height of one three-dimensional image can only be 20 cm, but larger CT and PET images of the patients were obtained by taking multiple three-dimensional images so that the bed holding the patient’s body is moved alongside the camera after each image. During the imagining, the patients lay on their back with their arms stretched above the head and the area from the top of the head to the upper thighs was scanned in 5–6 bed positions.
Because of the size differences of the patients, 13 patients had PET and CT images containing 341 transaxial slices, while the rest 65 patients had images of 395 transaxial slices. Each of these slices was of size \(256\times 256\) pixels in the PET images and \(512\times 512\) pixels in the CT images, and the voxel size was originally \(2.734\textrm{mm}\times 2.734\textrm{mm}\times 2.79\textrm{mm}\) for the PET images and \(0.98\textrm{mm}\times 0.98\textrm{mm}\times 2.79\textrm{mm}\) for the CT images. However, the PET and CT images were converted to two separate NIfTI files, both of which contained either \(512\times 512\times 341\) or \(512\times 512\times 395\) voxels of the size \(0.977\textrm{mm}\times 0.977\textrm{mm}\times 2.79\textrm{mm}\). A physician created three-dimensional binary masks of this same size to indicate all the voxels with cancerous cells in the primary tumor.
Transaxial slices depicting prostate cancer tumor were separated from the whole PET images with the help of the binary masks, these slices were converted into two-dimensional NumPy arrays of size \(512\times 512\), the borders of a fixed square [184 : 312, 148 : 276] were cropped out from each slice, and these \(128\times 128\) squares were scaled into a new size of \(64\times 64\) pixels. The location of this square is plotted to one transaxial slice from a CT image in Fig. 1, and as can be seen, it is between the bones of pubic symphysis and ischial tuberosity and contains the prostate. It was also checked from the binary masks that all their positive pixels are inside this square. The binary masks were then similarly cropped and scaled into the size of \(64\times 64\) pixels so that a pixel in the new mask was labeled positive if at least one of the corresponding four pixels in the original mask was positive. The numerical values of the pixels in PET slices were scaled onto the interval [0, 1] for each slice individually.
The final data contained 864 transaxial PET slices with 1–685 positive pixels per each slice, and these slices were from the 78 patients with 2–31 slices per patient. The data was divided into six sets of 13 different patients so that each set contained 145–149 slices or, equivalently, 16.8\(-\)17.2% of the data as shown in Table 1. In this way, we have six possible test sets with no overlap, and for each potential test set, we can set the contents of the five other sets into the training data. No information from the CT images was included in the final data.
Convolutional neural networks
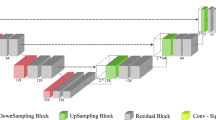
In this article, we use two CNNs that both follow the U-Net architecture introduced in Ronneberger et al. (2015). The first CNN used for typical image segmentation is the CNN built in Liedes et al. (2023), except it was modified to suit for images of \(64\times 64\) pixels. It has first a contracting path formed out of three sequences, each of which contains a convolution layer, a dropout layer, another convolution, and a pooling operation with ReLu activation functions. After the contracting path, the CNN has a similar expanding path, which has two sequences of three convolution operations, between of which there are a concatenation layer and a dropout layer. At the end of this CNN, there is a final convolution layer, but it has a linear function instead of the ReLu function. The architecture of this CNN is summarized in Table 2.
The second CNN used here to find suitable thresholds is the CNN designed in Hellström et al. (2023). This CNN consists of three pairs of convolutions, every one of which is followed by a pooling operation, and then there is a 3-layer feed forward after all these convolution-pooling sequences. The CNN uses a ReLu activation function on every layer except for the last one, whose ReLu function has been replaced here by a linear activation function. Table 3 shows the architecture of this second CNN.
The optimizer for both models was Adam (Adaptive Moment Optimization) with a learning rate of 0.001. The loss function was binary cross-entropy for the first CNN and mean squared error for the second CNN. The first CNN was trained on 30 epochs and the second one with 15 epochs, and during training of the CNNs, 30% of the training data was used for validation because these epoch numbers and this validation split produced the best segmentation results in the preliminary tests.
Evaluation of the models
For each predicted binary mask, we count the number of ground-truth positive pixels predicted correctly (TP), negative pixels incorrectly labeled as positive (FP), and positive pixels labeled as negative (FN), and then compute the Dice score defined as
Clearly, the greater the value of this coefficient is, the better the mask is, and its value of 1 means that each pixel was labeled correctly. To find the optimal thresholds required as the dependable variable for the second CNN, we created a function that chooses the two-digit threshold value between 0 and 1 that gives the maximum value of the Dice score when this threshold is used to convert data into a binary mask. Because the Dice scores do not follow the normal distribution, we consider the median and the interquartile range (IQR) for them instead of their mean values and standard deviation. The IQR is the difference between the 75th and 25th percentiles of the data, and it measures the spread of the data. For the same reason, we also use here the Wilcoxon signed-rank test to estimate whether the differences in the results of the three methods are statistically significant or not instead of the better-known Student’s t-test.
Structure of the experiment
First, we choose one of the six possible test sets, re-initialize the CNNs, and test all the three methods as follows: For the first method, we train the first CNN with the PET slices of the training set and their binary masks, compute the continuous predictions from the PET images of both the training and the test set, and find the constant threshold that gives the best median Dice score when used to convert all the predictions from the training set into binary masks and then use this same threshold to turn the predictions of the test set into binary masks. For the second method, we give our second CNN the PET slices of the training set with their optimal thresholds, train it, predict the results for the test set, and use the predicted thresholds to convert the PET slices of the test set into binary masks. For the third method, we first find the optimal thresholds for the output of the training set from the first CNN, train a re-initialized version of the second CNN by giving it the predicted training data and the found thresholds, predict the thresholds of the output of the test set from the first CNN, and use these predicted thresholds to convert this output into binary masks. The experiment is also described in the flowchart of Fig. 2. It is repeated for all the six sets.
Results
It was checked for the sake of comparison that if an optimal threshold was chosen for every PET slice separately, the median of the Dice scores would be 0.969 with an IQR of 0.0588. If the same two-digit threshold was used for converting all the PET slices, the best possible threshold would be 0.41, which would result in a median Dice score of 0.558 with an IQR of 0.457. For converting the continuous output of the first CNN to obtain the results of the first method based on the typical image segmentation, the thresholds 0.30, 0.19, 0.16, 0.31, 0.41, and 0.24 were used for the six test sets. Figure 3 also depicts the predicted values for the optimal thresholds to convert the PET slices directly into binary masks against the real values of these optimal thresholds for the first test set.
The predicted thresholds given by the second method to convert the PET slices into binary masks against the optimal thresholds, when the predictions are computed for the first test set by a CNN. The plot also contains the least squares regression line whose slope is 1.08 and intercept is \(-\)0.0611. Pearson’s correlation between these predicted and the optimal thresholds is 0.872, and the coefficient of determination \(R^2\) is 0.764
Table 4 contains the medians and the IQRs of the Dice scores, which were computed from the binary masks predicted with our three methods for the six different test sets separately. As we can see, there is much variation caused by the different training and testing data divisions, which is why this table has also the medians and the IQRs of the Dice scores of all the 864 predicted binary masks. When all the predictions are considered, we can see that the median of the Dice scores of the second method is the highest while the Dice scores of the third method have the lowest median. Furthermore, the third method produces better binary masks in those test sets that give the lowest values for the first method. There is not much difference in the IQRs between the methods or the test sets, but it must be noted that these values are very high.
The p-values of the Wilcoxon tests between the Dice scores of predictions given by the three methods are in Table 5. By comparing this table to Table 4, we notice that there is a statistically significant difference between the Dice scores of the first and the second method only at those times when the Dice scores of the second method have a higher median. The p-values for the Dice scores from the predictions of all the six sets also suggest that the second method is better than the first one in a statistically significant way. Furthermore, we see that the third method is also statistically worse than the two first ones.
To showcase results visually, Fig. 4 contains one transaxial PET slice in the first test set in Fig. 4A, its correct binary mask in Fig. 4B, the continuous output of the first CNN in Fig. 4C, this output converted into a binary mask with the constant threshold 0.30 used for the whole test set in Fig. 4D, the PET slice of Fig. 4A converted into a binary mask with a unique threshold 0.440 given by the second CNN in Fig. 4E, and the continuous output of Fig. 4C converted into a binary mask with a unique threshold 0.313 given by the second CNN in Fig. 4F. In other words, Fig. 4D, E, and F correspond with our first, second, and third methods. The Dice scores for the predicted binary masks in Fig. 4D, E, and F are 0.846, 0.789, and 0.846, respectively. The optimal threshold for converting for the PET slice of Fig. 4A is 0.50, which would have resulted in a fully correct binary mask. Additionally, Fig. 5 shows the four different segmentation results given by the second method with Dice scores varying between 0.366 and 0.851.
Discussion
According to our results, a CNN choosing a threshold for each transaxial PET slice separately to convert them into binary masks works better than a CNN performing the typical image segmentation when measured with the Dice scores of the predicted masks. The third method with a CNN finding optimal thresholds for the output of the usual image segmentation CNN performs worse than the two other methods, possibly because it has more potential causes of error. Still, if the CNN for typical image segmentation performs very poorly for some reason, its results could be potentially fixed by giving them the CNN that chooses unique thresholds to convert them to binary masks.
However, since the differences between the accuracy of these methods are relatively small, it is worth noting that using a CNN to find optimal thresholds directly for the PET slices is also considerably more computationally efficient. This is because the U-Net architecture is based on using a contracting and an expanding path so that the CNN can first see the whole picture and then focus on the details. If the U-Net CNN is built to choose suitable thresholds, it has less layers and it does not need to carry so many values over them because it only returns a single value for each image slice instead of a two-dimensional matrix with as many elements as pixels in the input. This makes the CNN faster, and because of its smaller number of weights, this new threshold optimization method requires less epochs for the CNN to converge, as noticed in the initial tests for this article.
One issue present with the predictions of all the three methods is that the segmentation is either very good or almost fully wrong. In other words, there is a high amount of variation in the results, as can be also seen in the IQRs of the Dice scores. This issue could be likely solved if there would be more data available to improve the training of the models, which would improve the medians of the Dice scores obtained. The methods studied here could be also potentially improved by using images with higher resolution than \(64\times 64\) pixels, and it could also be examined what happens if the pixel values of the PET slices are not scaled onto the interval [0, 1] or the slices are converted to colored images instead of using grayscale data. Adding anatomic information from multiparametric imaging could also improve results further. Namely, a CT/PET scan was performed for all the patients of this study, and while PET images give functional information about tissues, the CT images offer anatomical information about prostate size and location.
Another idea for the future study would be to design a CNN that can apply the threshold optimization method for three-dimensional segmentation of the prostate cancer tumor. Recently, Kostyszyn et al. (2021) obtained the Dice scores ranging 0.28–0.93 with median values of 0.81–0.84 with a three-dimensional U-Net CNN trained on PET images of 152 prostate cancer patients. Consequently, three-dimensional image segmentation gives more accurate results than two-dimensional segmentation, and our findings suggest that choosing thresholds performs better than regular image segmentation, so it would be interesting to see how well the three-dimensional version of the threshold optimization works. Furthermore, other possible topics of further research could be studying how changes in the PET image acquisition technique affect the results of segmentation.
The method of choosing unique thresholds with a CNN to perform segmentation could be also applied into other topics than the PET images of prostate cancer patients. One possible issue of this method is that it also labels such ground-truth negative regions outside of the prostate as positive if their pixel values are high enough, but this problem could be fixed with an algorithm removing the false positive segmentation. Also, by choosing a few different threshold values instead of one, this method could assist in the segmentation of colored images. Given its theoretically best results had a median Dice score closer to 97% here, this method certainly has much potential.
Conclusion
Three different methods of utilizing a CNN to perform intraprostatic tumor segmentation from 2D transaxial PET slices were studied. Out of these methods, the first one was typical image segmentation, the second one was using a CNN for choosing suitable thresholds for classifying pixel values in order to convert the images directly into binary masks, and the third one was a combination of the former two methods. The second and the third methods were fully new techniques introduced here for the first time. By analyzing the Dice scores of the predicted tumor masks, it was noticed that the second method performed the best. In fact, this new method works better than the typical image segmentation in a statistically significant way and is considerably faster and more computationally efficient.
Data availability
Patient data is not available due to ethical restrictions.
Code availability
Available at https://github.com/rklen/3_CNNs_for_PCa.
References
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Zheng X. TensorFlow: large-scale machine learning on heterogeneous systems. 2015
Anttinen M, Ettala O, Malaspina S, Jambor I, Sandell M, Kajander S, Rinta-Kiikka I, Schildt J, Saukko E, Rautio P, Timonen KL, Matikainen T, Noponen T, Saunavaara J, Löyttyniemi E, Taime P, Kemppainen J, Dean PB, Sequeiros RB, Aronen HJ, Seppänen M, Boström PJ. A prospective comparison of 18f-prostate-specific membrane antigen-1007 positron emission tomography computed tomography whole-body 1.5 T magnetic resonance imaging with diffusion-weighted imaging, and single-photon emission computed tomography/computed tomography with traditional imaging in primary distant metastasis staging of prostate cancer (PROSTAGE). Eur Urol Oncol. 2021;4(4):635–44.
Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS ONE. 2018;13(4):e0195798.
Chollet F, et al. Keras. GitHub. 2015
Guo Z, Li X, Huang H, Guo N, Li Q. Medical image segmentation based on multi-modal convolutional neural network: study on image fusion schemes. 2018 IEEE 15th Int Symp Biomed Imaging (ISBI 2018). 2018;903–07.
Global Cancer Observatory (GCO). Cancer today [Online analysis table]. 2022, January 11
Hellström H, Liedes J, Rainio O, Malaspina S, Kemppainen J, Klén R. Classification of head and neck cancer from PET images using convolutional neural networks. Sci Rep. 2023;13:10528.
Kostyszyn D, Fechter T, Bartl N, Grosu AL, Gratzke C, Sigle A, Mix M, Ruf J, Fassbender TF, Kiefer S, Bettermann AS, Nicolay NH, Spohn S, Kramer MU, Bronsert P, Guo H, Qiu X, Wang F, Henkenberens C, Werner RA, Baltas D, Meyer PT, Derlin T, Chen M, Zamboglou C. Intraprostatic tumor segmentation on PSMA PET images in patients with primary prostate cancer with a convolutional neural network. J Nucl Med. 2021;62(6):823–8.
Liedes J, Hellström H, Rainio O, Murtojärvi S, Malaspina S, Hirvonen J, Klén R, Kemppainen J. Automatic segmentation of head and neck cancer from PET-MRI data using deep learning. J Med Biol Eng. 2023. https://doi.org/10.1007/s40846-023-00818-8.
Malaspina S, Anttinen M, Taimen P, Jambor I, Sandell M, Rinta-Kiikka I, Kajander S, Schildt J, Saukko E, Noponen T, Saunavaara J, Dean PB, Sequeiros RB, Aronen HJ, Kemppainen J, Seppänen M, Boström PJ, Ettala O. Prospective comparison of 18 F-PSMA-1007 PET/CT, whole-body MRI and CT in primary nodal staging of unfavourable intermediate- and high-risk prostate cancer. Eur J Nucl Med Mol Imaging. 2021;48(9):2951–9.
Rainio O, Han C, Teuho J, Nesterov SV, Oikonen V, Piirola S, Laitinen T, Tättäläinen M, Knuuti J, Klén R. Carimas: an extensive medical imaging data processing tool for research. J Digit Imaging. 2023
Rawla P. Epidemiology of prostate cancer. World J Gastroenterol. 2019;10(2):63–89.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer: Cham; 2015. pp. 234–241
van Rossum G, Drake FL. Python 3 reference manual. CreateSpace. 2009
Sciarra A, Gentilucci A, Salciccia S, von Heland M, Ricciuti GP, Marzio V, Pierella F, Musio D, Tombolini V, Frantellizzi V, Pasquini M, Maraone A, Guandalini A, Maggi M. Psychological and functional effect of different primary treatments for prostate cancer: a comparative prospective analysis. Urol Oncol Semin Orig Investig. 2018;36(7):340.e7-340.e21.
Sharif MS, Abbod M, Amira A, Zaidi H. Artificial neural network-based system for PET volume segmentation. Int J Biomed Imaging. 2010;2010: 105610.
Townsend DW. Physical principles and technology of clinical PET imaging. Ann Acad Med Singap. 2004;33(2):133–45.
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Jarrod Millman K, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17(3):261–272.
Funding
Open Access funding provided by University of Turku (including Turku University Central Hospital). The first author was financially supported by the Finnish Culture Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rainio, O., Lahti, J., Anttinen, M. et al. New method of using a convolutional neural network for 2D intraprostatic tumor segmentation from PET images. Res. Biomed. Eng. 39, 905–913 (2023). https://doi.org/10.1007/s42600-023-00314-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-023-00314-7