Abstract
Introduction
Magnetic resonance imaging (MRI) is the most used medical modality for diagnosis and monitoring of multiple sclerosis (MS). A segmentation process is an important task to quantify lesion and its progression. However, manual segmentation of 3D images is tedious, time-consuming, and often not reproducible. The state of the art presents results with room for improvements. Consequently, a semiautomatic segmentation process is proposed and described in this study.
Methods
The method consists on a 3D segmentation semiautomatic process for MS lesions in MRI. It initiates by firstly carrying out a preprocessing stage; thus, contrast adjustment is applied to enhance sclerosis regions from other brain information. Secondly, a feature extraction block based on fuzzy connectedness is performed so as to isolate sclerosis lesions from other brain regions. Finally, 3D brain reconstruction is executed along with sclerosis to provide a useful 3D information.
Results
The robustness of this approach is demonstrated by high correlation between the results and their corresponding gold standard. The results were also obtained by computing parameters of accuracy of image segmentation, as well as overlap Dice. The proposed method reached true positive of 75.61%, false positive of 16.37%, and DICE of 78.23%.
Conclusion
The high correlation between specialist and proposed approach outcome, a better monitoring of the disease, is provided; the specialist can understand the patient’s symptoms, thereby increasing the patient’s quality of life.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is the major cause of non-traumatic neurological disability in young adults in Europe and North America, affecting approximately 2.5 million people worldwide. It is an autoimmune disease of the central nervous system (CNS), in which inflammatory demyelination of axons causes focal lesions in CNS white matter (WM) (Roy et al. 2018; Tomas-Fernandez and Warfield 2015). Although the cause of MS is still unknown, several studies suggest it may be caused by combination among genetic, environmental, and immunological factors (Tomas-Fernandez and Warfield 2015).
MS can present inflammatory and neurodegenerative components. The acute demyelination and inflammatory axonal transection may be responsible for the disease symptoms. Degeneration appears to be the main reason for the disability and progression in MS. Several putative therapeutic strategies for remyelination and neuroprotection are now transitioning from the laboratory to early phase clinical trials (Bhargava et al. 2015).
Nowadays, magnetic resonance imaging (MRI) is used in the diagnosis and monitoring of MS that is because the sensitivity of structural MRI shows WM lesions in time and space without contrast injection (Roy et al. 2018; Valverde et al. 2017; Jain et al. 2015). These WM lesions are visible on a MRI brain scan and appear hyperintense on T2-weighted or fluid-attenuated inversion recovery (FLAIR) images.
Since MRI was introduced in the early 1980s to diagnose and assess MS, this exam has become the primary imaging modality to monitor its natural evolution (Tomas-Fernandez and Warfield 2015). Assessment of the disease implication using MRI for research and clinical trials requires quantification of the volume on images. It has been shown that volume and location of lesions by their segmentations are important biomarkers of MS and can be used to detect its onset or track its progression. However, lesions vary in size, shape, intensity, and location, which make their automatic and accurate segmentation challenging (Dimitrova 1977).
Manual delineations are considered the gold standard (GS), but segmenting lesions from 3D images is tedious, time-consuming, and often not reproducible (Jain et al. 2015; Roy et al. 2018). Besides that, errors can occur due to low lesion contrast and unclear boundaries caused by changing tissue properties and partial volume effects (Tomas-Fernandez and Warfield 2015). Furthermore, there is an inherent reliability challenge associated with lesion segmentation. Images produced by any imaging device are inherently fuzzy. This fuzziness comes from several sources like spatial and temporal resolution limitations, blur and/or noise, and background intensity variation. In addition to these factors, different tissues, organs, and anatomic structures manifest heterogeneity of intensity values of object regions in acquired images (Udupa and Saha 2003). This problem is accentuated in MRI, because this type of exam does not have any uniform intensity scale (like computed tomography); acquisition of images in different scanners and with different contrast properties can add complexity to the segmentation (Roy et al. 2018).
A semiautomatic or automatic MS segmentation method is required to reduce the time of this task and intra-rater variability uncertainty among segmentation made by different specialists (Egger et al. 2017; Udupa et al. 1997). This will be especially important for large clinical trials, since many images need to be analyzed and processed. For clinical practice, this segmentation method would allow the measurement of lesion volumes, standardizing and quantifying MRI observation (Jain et al. 2015).
Significant works on MS segmentation have been published in recent years (Beaumont et al. 2016; Egger et al. 2017; Roura et al. 2015). In a study by Beaumont et al. (2016) is presented an automatic segmentation from multimodal graph cutting. The results of this segmentation are good, but they vary because they depend on the input parameters of the algorithm, which are directly associated with the total load of the lesion. The automation of this method is complicated because the initialization of the parameters is very important to achieve satisfactory results. Egger et al. (2017) evaluated the Schmidt et al. (2012) algorithm in an independent dataset and compared the results with data from three experienced raters. In this work, they determine the MS lesion using two algorithms that runs under the two statistical parametric mapping (SPM) packages: Lesion growth algorithm based (LGA) on SPM8, LGA on SPM12, and lesion predicting algorithm (LPA) based on SPM12. In this evaluation, they found a strong correlation between manual and automated segmentation, obtaining DICE 0.6, 0.53, and 0.57 for LGA SPM8, LGA SPM12, and LPA SPM12, respectively. In Roura et al. (2015), an automated segmentation using T1w and FLAIR images is explored. This approach consists of two steps: segmentation of the brain tissue according to gray matter, white matter, and cerebrospinal fluid in T1w images, followed by the segmentation of the lesions as outliers to the brain tissue in the FLAIR image. In the study of Roura et al. (2015), the quantitative evaluation reached DICE of 0.3, 0.33, and 0.43 in three distinct databases.
These automatic targeting methods prevent user variability and reduce time consumption, but the accuracy of these methods has not yet achieved their highest possible potential. This feature is less pronounced in semiautomatic segmentations that require a specialist to initialize. In the literature, there is still no viable standard tool for daily clinical practice. Automatic detection of multiple sclerosis lesions is still a challenging problem, living room for, in a primary step, focus on a reliable semiautomatic method.
In this paper, a semiautomatic segmentation based in fuzzy connectedness (FC) was introduced as a robust method for WM lesion segmentation on 3D FLAIR MRI. The method is independent of scanner and acquisition protocol and also does not require a huge large training image database of expert lesion segmentations. Consequently, the objective of this study is to construct and validate a proposed segmentation method for a 3D segmentation, using as core operation fuzzy connectedness with fixed weights to compute the affinity level between pixels. In addition, the evaluation was performed in a set of MRI scans to compute its accuracy; hence, results were compared quantitatively and qualitatively with GS made manually by specialists. The applied parameters of accuracy were extracted from the proposed approach: a framework for evaluating image segmentation algorithms (Udupa et al. 2006) and overlap Dice (Dice 1945); thus, punctual comparison with the results obtained with the literature was allowed.
The project is running under a strong collaboration of complementary work teams from Federal University of São Paulo (UNIFESP), specifically, between the Medical Imaging Processing Group of Institute of Science and Technology (ICT-UNIFESP) coordinated by Prof. Dr. Matheus Cardoso Moraes and the group of DDI-UNIFESP coordinated by Prof. Dr. Nitamar Abdala.
Material and methods
In this study, a semiautomatic segmentation of MS areas was constructed and evaluated in a set of 32 MS lesions in different MRI exams made by scanners Philips Achieva 3TX and Siemens Skyra 3TX. Each exam contains about 143 slices in DICOM-format, 16-bits resolution, with positive values. The exams were provided by the Department of Imaging Diagnosis of the São Paulo School of Medicine (DDI-UNIFESP) through XNAT platform: PACS Research Management and storage of data and clinical images of research projects in DDI-UNIFESP. The ethics committee with the number 03830718.9.0000.5505 approved the study protocol to allow the medical images manipulation.
The evaluation was performed by computing the mean and standard deviation of true positive (TP), false positive (FP), false negative (FN) (Udupa et al. 2006), overlap (OR) (Kupinski and Giger 1998), and overlap Dice (OD) (Dice 1945) was also calculated to compare the results obtained previous work. Experts manually made and revised the gold standards under the collaboration described above.
Three main stages are applied to describe the segmentation methodology. The contrast adjustment is performed to intensify the pixels of the region with MS during preprocessing. The feature extraction is firstly performed by acquiring information from the region of interest (ROI) in three dimensions by constructing a connectivity map using fuzzy connectedness methodology (Udupa and Samarasekera 1996). It is followed by post-processing using binarization, and mathematical morphology takes place to enhance the previously extracted information. In the final step, the 3D reconstruction of brain volume with the segmented MS is carried out, hence providing better visualization of the brain regions affected by MS. The block diagram shown in Fig. 1 resumes the segmentation process.
Stage 01 ➔ preprocessing
This stage is divided in two steps: brain isolation and contrast adjustment. Medical images are characterized by a composition of small differences in signal intensities between different types of tissues, noise, manufacture, etc. Hence, differences, ambiguities, and uncertainties are, by default, introduced during image formation. These imprecisions could make difficult a thorough discrimination of the exact location and area of the ROI (Pednekar and Kakadiaris 2006). Because of these circumstances, an intensity level normalization process takes place by contrast enhancement, hence normalizing and minimizing possible difference among scanners, providing an intensity levels normalization concerning sclerosis’ intensities and surrounding.
Since MS is better observed in T2 and FLAIR images as a region with high-intensity pixels, the contrast of the image is adjusted to enhance the demyelinated regions. And to normalize the input images, it is needed to preprocess the images with filters and contrast adjustments. This makes the method robust and independent of the sensor used.
Figure 2 displays the input and output of this stage with one slice from an MRI exam utilized in this work.
Step 01 ➔ brain isolation
At the beginning of this step, the original image Io (Fig. 3a ) is the input. First, from Io an image with contrast elongation Iac at the lower threshold of 0.15 and greater than 0.2 was created (Chang and Wu 1998). Second, an image with an equalized histogram Ihe was generated from Io. Consequently, we multiplied these two results, Iac by Ihe, and applied Otsu binarization process, obtaining Iotsu (Otsu 1979). With Iotsu, an opening operation was carried out with a sphere of radius 6 voxels to isolate the brain, which is the ROI, from other regions and information included in the image. The brain Ibrain (Fig. 3b) was located as the binarized region with the largest volume. This step was carried out with the purpose of improving the next step of contrast adjustment of the original image, prioritizing the pixels only pertinent to the cerebral volume of the exam.
Results of each preprocessing calculation. a Original image, Io. b Binarized isolated brain, Ibrain. c Isolated brain with original image values, Ibo. d Ibo with Gaussian filtering, Ifilt. e Shade flat field correction, Iflat. f Edge sharpening, Isharp. g Preprocessing final image with histogram adjust, Ia
Step 02 ➔ contrast adjustment
Firstly, Io and Ibrain were multiplied, isolating only brain information, resulting in the brain with values of the original image Ibo (Fig. 3c). After that, we applied a spatial Gaussian filter obtaining Ifilt (Fig. 3d), the parameters of filtering were kernel of 9 × 9 pixels, and the mean of the local intensities is covered by the kernel and standard deviation (sigma) of 0.8. Next, a flat-field correction (Seibert et al. 1998), resulting in Iflat followed by an edge sharpening obtaining Isharp was serially performed to reduce shading distortion (Fig. 3e and f ). Finally, the image histogram was adjusted with a narrowing at 0.2 and 0.7 thresholds resulting Ia (Fig. 3g).
The used parameters’ values were found empirical, by firstly visually evaluating the resultant value that mainly emphasizes the ROI. The number of trying and spend time during this analytical/empirical process was not measured.
Stage 02 ➔ Feature Extraction
This stage combines operations to acquire and polish the most of MS information. First, fuzzy connectedness methodology (Udupa and Samarasekera 1996; Udupa et al. 1997) is applied to increase discrimination between MS tissues and the rest of image. Secondly, a polishing and enhancement of the discriminated information are carried out by a binarization and mathematical morphology process.
Step 01 → fuzzy connectedness is a semiautomatic segmentation method based on region growing. The process relies on a combination of criteria that takes into account homogeneity and intensity features from a selected region (Cardenas et al. 2013). The fuzzy connectedness process starts with a MS ROI selected and a seed defined (initialization). Then, a voxel of a ROI must be selected by user, as it is a semiautomatic method. Second, the image homogeneity and intensity features are combined among a seed and its neighbors in a parameter called affinity (Udupa and Samarasekera 1996). Thirdly, by using the Dijkstra graph theory methodology, the connectivity map among the planted seed and each voxel of the image is finally constructed. Please, for more details about fuzzy connectedness algorithm, refer to (Nyúl et al. 2002; Udupa and Samarasekera 1996; Wilcox and Hirshkowitz 2015).
During the initialization process, a pixel of Ia ROI should be selected (first seed). With the selected pixel, the mean and standard deviation of the local homogeneity (m1 and s1) and intensity (m2 and s2) of the objects were calculated (Cardenas et al. 2013; Nyúl et al. 2002; Udupa and Samarasekera 1996; Wilcox and Hirshkowitz 2015). Specifically, for this approach, the ROI was acquired by choosing a slice containing a MS region and clicking in this ROI’s central voxel to acquire the required intensity information in a window of 15 by 15 pixels over the slice’s ROI. We chose a 15 by 15 window size, since it showed to be sufficient to acquire the information and is sufficiently small to not overcome ROI regions.
Once this is done, the affinity of the first seed with its six neighbors (north, south, east, west, back, and front) was calculated. The highest affinity values of this first interaction were used as reference for a stop condition criterion, proposed for this application to avoid computing the complete image Connectivity, decreasing computational cost. The established value was empirically defined as 70% of the highest affinity. Hence, the region growing is performed until the connectivity drops to the mentioned value.
While the stopping condition is not reached, a growth loop (GL) process is still running. This process provides the intrinsic fuzzy connectedness interactions (Cardenas et al. 2013; Nyúl et al. 2002; Udupa and Samarasekera 1996; Wilcox and Hirshkowitz 2015). Hence, first we compute the homogeneity μψ(c, d) and intensity μϕ(c, d) between the current seed (c) and the analyzed pixel (d), computed through:
in which f(c) and f(d) are respectively the values of the image at the position of the seed pixel and the neighbor pixel analyzed. With these two parameters, GL computes the similarity level μɑ(c,d) between analyzed pixel and current seed, denominated affinity, and determined by:
where w1 and w2 are respectively the weights assigned to homogeneity and intensity with values of 0.3 and 0.7. Values were calibrated and chosen by focus on the best segmentation result during a parameter calibration stage.
Consequently, the connectivity (pixel pertinence level in the ROI) of the analyzed pixel μk(d) is updated as the minimum between μɑ(c,d) and the connectivity of the current seed μk(c) from:
with Eq. 4, the affinity value is not able to increase, continuing the same value of seed connectivity or decreasing until reaching the stop condition. As mentioned above, this is carried out for the 6 neighbors of the current seed. If the neighbor was already assigned previously as a seed, we call it an ex-seed; hence, the calculations are not performed for that pixel, since, by being a seed, it was already classified as part of the desired object. The neighbors that were analyzed are added as new seeds in a queue of seeds and sorted according to connectivity. The seed that was the current one is removed from this queue and placed an indicator of ex-seed. The dynamic of the process considers the pixel that has the highest current connectivity as the new seed.
Upon reaching the stop condition, GL is terminated, and we have the 3D connectivity matrix (Mc) with values between 0 and 1 (Fig. 4 Mc) . The stop condition was designed and calibrate to terminated as soon as the region growing process leaves the MS area. As mentioned above, it assures the extraction of the lesion and enormously decreases computational cost.
Step 02 ➔ post-processing
In this step, Mc is binarized (Ob) without threshold, since the stop condition makes Mc, contains only MS information all values is transformed to 1 resulting in (Fig. 4Ob). Next, with the binary object, we perform mathematical morphology (Haralick and Sternberg 1987) closing operation in Ob to fill any small apertures caused by noise resulting in Of (Fig. 4Of). Figure 4 demonstrates a one slice process; nevertheless, the procedure is occurring in a 3D space, since front and back voxels were considered during connectivity computation.
(b) Connectivity matrix resulting from FC. (c) Binarized object. (d) Object after closing operation
Stage 03 ➔ 3D reconstruction
The brain 3D reconstruction is performed to facilitate visualization of the brain volumes affected by MS, improving medical analysis, and enormously increase the success of clinical decisions. Accordingly, in an accurate and overall view, the specialists will be able to take advantage of special details regarding the MS volume and location, leading to a better understanding of the symptoms that the patient presents or will present. In addition, with the follow-up through examinations, it will be possible to observe the volumetric growth of the disease.
In this step, we need the MS extracted Of, (Fig. 5) which was described above and the complete brain isolated Ibrain (Fig. 5) computed in preprocessing stage. With DICOM voxel information, a interpolation process is performed, and the brain volume is rebuilt along with segmented MS, Of (Paluszek and Thomas 2017).
Results
To evaluate the method outcomes, the proposed method was applied in 32 MS lesions from FLAIR MRI exams of patients with MS performed on the Philips Achieva 3TX and Siemens Skyra 3TX, containing about 143 slices by exam. The exams were provided by DDI-UNIFESP XNAT platform: PACS Research Management and storage of data and clinical images of research projects. The computational cost was based on a computer with an Intel® Core ™ i7-4790 K CPU @ 4.00GHz processor, 16GB RAM, Windows 10 64-bit and MATLAB 2016a software from the Image and Signal Processing Laboratory without any code optimization. The MRI exams have been segmented and compared with GS made by experts from DDI-UNIFESP.
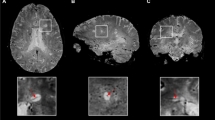
In Figs. 6 and 7, some results of the proposed method are displayed. Figure 6 shows that the corresponding boundaries of segmented lesions by the proposed method can reach ROI boundaries. However, some pixels with lower intensity (seen with gray color in figure), excluded by the proposed method as part of the lesion, can, in fact, belong to the lesion. In Fig. 7, the high similarity between the images segmented by this approach and their GS is noticeable.
Results obtained with the proposed method. In the left side the contours of the GS are displayed, the contour of the segmentation with the proposed method is in the middle, and on the right is exposed the 3D reconstruction of the brain with the MS lesion (in green the lesion segmented by specialist and in brown by the proposed method)
The statistical evaluation was carried out by applying the proposed methodology in the 32 MS lesion volumes. Next, each segmented result was compared with its corresponding gold standard (GS) made manually by an expert. The numerical assessment of accuracy was obtained by computing corresponding parameters of accuracy true positive (TP), false positive (FP), and false negative (FN) (Udupa et al. 2006), as well as overlap (Kupinski and Giger 1998) and overlap Dice (Dice 1945). The parameter of all 32 volumes is shown in Table 1, and the mean and standard deviation is shown in Table 2.
As can be observed in Table 1, the results obtained have high accuracy with TP around 75% and an FP near 16%. FN is complementary to TP. The high FN value means the voxel that the method did not register as belonging to the lesion. This can occur due to the low sensitivity in the method of weights applied empirically or also by the value of the stop condition to decrease or use the computational value.
To compare with other results, we calculated the DICE which was 78.23%. Dice similarity coefficient values range from 0 to 1, where 0 corresponds to no overlap between two objects and 1 corresponds to perfect overlap. The false-positive fraction and true-positive fraction were computed for each lesion (Fig. 8) to indicate the percentage of voxels correctly or incorrectly classified as lesion by the method.
The average segmentation time of all lesions was 2.97 s ± 0.331 s, much faster than manual targets that can last for minutes. This manual segmentation was performed by the specialist collaborator of this project. This result can be improved by code optimization and/or by using different programming language such as C++ or Python.
In this graph, we can see that in most of the lesions, the TP was above average, and the FP was below average. The lesions that do not obey this rule can be for reasons such as possible imprecision of the specialist segmentation moment in regions that are not intense but are homogeneous, and as our method is mathematical, it is possible to carefully evaluate the relevance of the pixel in ROI. And small lesions that have a reduced area for initialization of segmentation may not present adequate values of intensity and/or homogeneity, so that the method may not ideally initialize, compromising targets, neighboring pixels of the first seed.
Discussion
The use of MRI to diagnose demyelinating diseases makes it necessary to develop computational methods that assist the specialist (Roy et al. 2018; Storelli et al. 2016). However, despite efforts, lacking accurate methods and results makes the task of MS segmentation challenging.
This research was carried out to evaluate the performance of fuzzy connectedness in the segmentation of MS in MRI, seeing that a non-manual segmentation method is required for this task. The developed method is working fast, saving the specialist’s time when performing MS segmentation. The obtained results, computed in 3D domain with challenge images, also indicate a relatively high correlation with manual segmentation from specialists, making the follow-up of the disease less susceptible to subjective interpretation of the different specialists.
The methodology presented in this work for the segmentation of MS in the brain was divided into three stages. The first, preprocessing, the contrast of the image is adjusted in order to evidence the ROI. The second, feature extraction, uses fuzzy connectedness to calculate the connectivity matrix for ROI, binarization, and mathematical morphology. Finally, the brain with MS is reconstructed in three dimensions to obtain volumetric visualization of the regions affected by the disease.
The high TP value and low FP value indicate that this segmentation method has a smaller variation comparing with the segmentations performed by a specialist or different specialists (Udupa et al. 1997). Moreover, taking into account the difficulties of the images because they are from different sources, the accuracy and robustness of the method are verified, thus offering a new semiautomatic alternative to carry out the segmentation of one of most common demyelinating diseases.
Concerning previous work in the literature, a thorough comparison of outcomes from different works is beyond the scope of this study, since the datasets, evaluation indexes, and computational resources are different. Nevertheless, compare and contrast corresponding outcome are important to help corroborate efficiency. The work produced by Jain and colleagues (Jain et al. 2015) compare 3 unsupervised classification methods of automatic segmentation based on stochastic modeling of voxel intensity distribution Msmetrix, LST, and Lesion-TOADS with Dice 0.69 ± 0.14, 0.71 ± 0.18, and 0.63 ± 0.17, respectively. Although it is tempting to propose automatic segmentation, these methods of unsupervised classification do not yet present high accuracy. In comparison, in our proposed approach, the proposed method reached DICE of 78.23 ± 8.51 with a semiautomatic segmentation method. According to the literature in this field, recent techniques proposed for MS lesion segmentation include supervised learning methods such as decision random forests, ensemble methods, non-local means, and k-nearest neighbors (Valverde et al. 2017). This type of classification has good results in the training dataset but contains disadvantages such as the construction of a considerably large training dataset that encompasses MS lesions of all possible forms, intensities, and heterogeneous textures in WM (Jain et al. 2015).
Because it is semiautomatic, this method requires the specialist to identify the area with sclerosis and initialize the segmentation. In lesions of medium and large region, the method is very robust and fast. A not so high accuracy when small sclerosis regions are concerning may be seen as a limitation, as the method may end up computing regions that do not belong to the sclerosis that is because the mean and standard deviation of the region of interest may not be representative enough for small regions. Another possible reasons are the empirical values found in the initiation and applied filters. Thus, inaccurately initializing the segmentation process may limit the possible best outcome. Moreover, if a region without MS is selected, through the affinity among voxels, it will be wrongly segmented; hence, the correct choice of specialist is important.
Conclusion
The methodology developed and applied in this study presented high TP and low FP values. The segmentation time, 2.97 s ± 0.331 s, comparing with the manual, is much faster than manual ones, in which can last for minutes; in addition, manual segmentation may become a hard and time-consuming task depending on the dataset size. Consequently, with this study, it was possible to observe the robustness of fuzzy connectedness in the segmentation of multiple sclerosis, using simple weights to calculate the affinity between voxels. The main contributions of this work were (i) a preprocessing stage to enhance MS areas and volumes; (ii) the specific weight’s values to improve fuzzy connectedness in MS segmentation and evaluation; (iii) a set of mathematical morphology operations to reconstruct a binary version of MS volume; and (iv) a 3D reconstructed brain with MS regions segmented and highlighted, so as to improve clinical analysis allowing the specialist to observe encephalon affected by the disease.
In order to overcome the limitations mentioned at the end of the Discussion section, as well as increasing accuracy, future works will focus on increasing dataset to be able to associate deep and/or machine learning methods for this application. Consequently, in future work, we will investigate the potential of Bhattacharyya affinity and dynamic weight functions for FC allied to CNN to make segmentation more reliable, overcoming initialization dependence, making it completely automatic, and improving accuracy (Roy et al. 2018; Valverde et al. 2017). We also can create a machine learning to find the best parameters for weights, filters, and windows size to start the method.
References
Beaumont, J. et al. Multiple sclerosis lesion segmentation using an automated multimodal graph cut. HAL, 2016.
Bhargava P, Lang A, al-Louzi O, Carass A, Prince J, Calabresi PA, et al. Applying an open-source segmentation algorithm to different oct devices in multiple sclerosis patients and healthy controls: implications for clinical trials. Mult Scler Int. 2015;2015. ISBN: 2090-2654, ISSN: 20902662:1–10. https://doi.org/10.1155/2015/136295.
Cardenas DAC, Moraes MC, Furuie SS. Segmentação do lúmen em imagens de IOCT usando fuzzy connectedness e reconstrução binária morfológica. Rev Brasil Engenharia Biomed. 2013. ISSN: 15173151;29(1):32–44. https://doi.org/10.4322/rbeb.2013.004.
Chang D, Wu W. Image contrast enhancement based on a histogram transformation of local standard deviation. 1998;17(4):518–531.
Dice LR. Measures of the amount of ecologic association between species author. Ecology. 1945;26(3):297–302. https://doi.org/10.2307/1932409.
Dimitrova E. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. Akusherstvo i Ginekologiya. 1977. ISBN: 1558-254, ISSN: 1558254X;16(6):452–7. https://doi.org/10.1109/TMI.2016.2528821.
Egger C, et al. NeuroImage: clinical MRI FLAIR lesion segmentation in multiple sclerosis: does automated segmentation hold up with manual annotation? NeuroImage: Clin. 2017. ISSN: 2213-1582;13:264–70. https://doi.org/10.1016/j.nicl.2016.11.020.
Haralick, R. M.; Sternberg, S R Image analysis morphology 1987;4:532–550.
Jain S, et al. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage: Clin. 2015. ISBN: 1460-9568, ISSN: 22131582;8:367–75. https://doi.org/10.1016/j.nicl.2015.05.003.
Kupinski MA, Giger ML. Automated seeded lesion segmentation on digital mammograms. IEEE Trans Med Imaging. 1998;17(4):510–7.
Nyúl LG, Falcão AX, Udupa JK. Fuzzy-connected 3D image segmentation at interactive speeds. Graph Model. 2002. ISSN: 15240703;64(5):259–81. https://doi.org/10.1016/S1077-3169(02)00005-9.
Otsu N. A threshold selection method from gray-level histograms. IEEE Transact Syst Man Cybern. 1979;9(1):62–6.
Paluszek, M.; Thomas, S. MATLAB graphics. In: MATLAB Machine Learning, 2017. ISBN: 9781484222492.
Pednekar AS, Kakadiaris IA. Image segmentation based on fuzzy connectedness using dynamic weights. IEEE Trans Image Process. 2006. ISBN: 1057-7149, ISSN: 10577149;15(6):1555–62. https://doi.org/10.1109/TIP.2006.871165.
Roura E, et al. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015. ISBN: 0023401515522;57:1031–43. https://doi.org/10.1007/s00234-015-1552-2.
Roy, S. et al. Multiple sclerosis lesion segmentation from brain MRI via fully convolutional Neural Networks no 2013, 2018.
Seibert JA, Boone JM, Lindfors KK. Flat-field correction technique for digital detectors, Proc. SPIE 3336, Medical Imaging: Physics of Medical Imaging, 1998.
Storelli L, Pagani E, Rocca MA, Horsfield MA, Gallo A, Bisecco A, et al. A semiautomatic method for multiple sclerosis lesion segmentation on Dual-Echo MR imaging: application in a multicenter context. Am J Neuroradiol. 2016. ISBN: 0195-6108, ISSN: 1936959X;37(11):2043–9. https://doi.org/10.3174/ajnr.A4874.
Tomas-Fernandez X, Warfield SK. A model of population and subject (MOPS) intensities with application to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2015. ISBN: 0278-0062 VO - 34, ISSN: 1558254X;34(6):1349–61. https://doi.org/10.1109/TMI.2015.2393853.
Udupa JK, Saha PK. Fuzzy connectedness and image segmentation. Proc IEEE. 2003. ISBN: 0018-9219, ISSN: 00189219;91(10):1649–69. https://doi.org/10.1109/JPROC.2003.817883.
Udupa JK, Samarasekera S. Fuzzy connectedness and object definition: theory, algorithms, and applications in image segmentation. Graphical Models Image Process. 1996;58(3):246–61.
Udupa JK, Wei L, Samarasekera S, Miki Y, van Buchem MA, Grossman RI. Multiple sclerosis lesion quantification using fuzzy- connectedness principles. IEEE Trans Med Imaging. 1997. ISBN: 0278-0062, ISSN: 02780062;16(5):598–609. https://doi.org/10.1109/42.640750.
Udupa JK, LeBlanc VR, Zhuge Y, Imielinska C, Schmidt H, Currie LM, et al. A framework for evaluating image segmentation algorithms. Comput Med Imaging Graph. 2006. ISBN: 0895-6111, ISSN: 08956111;30(2):75–87. https://doi.org/10.1016/j.compmedimag.2005.12.001.
Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova JC, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017. ISBN: 1095-9572 (electronic) 1053-8119 (linking), ISSN: 10959572;155:159–68. https://doi.org/10.1016/j.neuroimage.2017.04.034.
Wilcox T, Hirshkowitz A. Iterative relative fuzzy connectedness for multiple objects with multiple seeds. Comput Vis Image Underst. 2015;85(01):1–27. https://doi.org/10.1016/j.neuroimage.2013.08.045.
Schmidt et al., 2012. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage. 2012;59:3774–83.
Acknowledgments
Laboratory of Image and Signal Processing of the Institute of Science and Technology (LaPIS- ICT-UNIFESP) and Department of Diagnostic Imaging (DDI-UNIFESP-SP) for the provision of MRI with the XNAT system and Coordination for the Improvement of Higher Education Personnel (CAPES) for financial support. .
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have conflict of interest.
Ethical approval
University Ethics Committee approved the study (approval number 3.243.081)
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Arruda, A.L.C., Vital, D.A., Kitamura, F.C. et al. Multiple sclerosis segmentation method in magnetic resonance imaging using fuzzy connectedness, binarization, mathematical morphology, and 3D reconstruction. Res. Biomed. Eng. 36, 291–301 (2020). https://doi.org/10.1007/s42600-020-00070-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-020-00070-y