Abstract
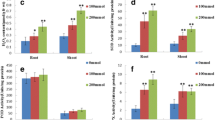
Biochemical studies show a definite relation of salinity stress with plant proteome alteration. The impact of salinity stress on the root proteins is elucidated here. The experiment included two genotypes of wheat, KH-65 and PBW-373; this experiment was tested against 0 and 300 mM NaCl for 48 h. Liquid chromatography combined with mass spectrometry (LC–MS/MS) was employed to analyze extracted proteins. Significant results were obtained regarding the rarely high number of proteins, peptides, and protein groups as the chromatograms of two genotypes revealed as many as 21,863 proteins, 5133 protein groups, and 27,881 peptides. Analysis of genotype-specific variations shows that change in the expression pattern of proteins corresponds with different levels of salinity tolerance as seen through the overwhelming higher up-regulation in tolerant genotype (KH-65) and comparative down-regulation of proteins in susceptible genotype (PBW-373). Some of such proteins, auxin-responsive protein, peroxidase and vacuolar protein sorting-associated protein 41 homolog, are mentioned.



Similar content being viewed by others
References
Ahn HK, Kang YW, Lim HM, Hwang I, Pai HS (2015) Physiological functions of the COPI complex in higher plants. Mol Cells 38(10):866
Baral A, Shruth KS, Mathew MK (2015) Vesicular trafficking and salinity responses in plants. Iubmb Life 67(9):677–686
Bilkis A, Islam MR, Hafiz MHR, Hasan MA (2016) Effect of NaCl induced salinity on some physiological and agronomic traits of wheat. Pak J Bot 48:455–460 ((Corpus ID: 189805866))
Capriotti AL, Borrelli GM, Colapicchioni V, Papa R, Piovesana S, Samperi R, Stampachiacchiere S, Laganà A (2014) Proteomic study of a tolerant genotype of durum wheat under salt-stress conditions. Anal Bioanal Chem 406(5):1423–1435
Chen J, Cheng T, Wang P, Liu W, Xiao J, Yang Y, Hu X, Jiang Z, Zhang S, Shi J (2012) Salinity-induced changes in protein expression in the halophytic plant Nitraria sphaerocarpa. J Proteom 75(17):5226–5243
Dejonghe W, Sharma I, Denoo B, De Munck S, Lu Q, Mishev K, Bulut H, Mylle E, De Rycke R, Vasileva M, Savatin DV (2019) Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat Chem Biol 15(6):641–649
FAO (2021) FAO soils portal, salt affected soils. http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/salt-affected-soils/more-information-onsalt-affected-soils/en/
Faurobert M, Pelpoir E, Chaïb J (2007) Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. In: Thiellement H, Zivy M, Damerval C, Méchin V (eds) Plant proteomics: methods and protocols. Methods in molecular biology, vol 335. Humana Press Inc., Totowa, New Jersey, United States, pp 9–14.
Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS (2011) The fifth adaptor protein complex. PLoS Biol 9(10):e1001170
Hussain S, Zhu C, Bai Z, Huang J, Zhu L, Cao X, Nanda S, Hussain S, Riaz A, Liang Q, Wang L (2019) iTRAQ-based protein profiling and biochemical analysis of two contrasting rice genotypes revealed their differential responses to salt stress. Int J Mol Sci 20(3):547
Jones JB Jr (1982) Hydroponics: its history and use in plant nutrition studies. J Plant Nutr 5(8):1003–1030
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13(4):889–905
Kim SJ, Brandizzi F (2012) News and views into the SNARE complexity in Arabidopsis. Front Plant Sci 3:28
Kubala MH, Norwood SJ, Gomez GA, Jones A, Johnston W, Yap AS, Mureev S, Alexandrov K (2015) Mammalian farnesyltransferase α subunit regulates vacuolar protein sorting-associated protein 4A (Vps4A)-dependent intracellular trafficking through recycling endosomes. Biochem Biophys Res Commun 468(4):580–586
Marjamaa K, Kukkola E, Lundell T, Karhunen P, Saranpaa P, Monolignol FKV (2006) Oxidation by xylem peroxidase isoforms of Norway spruce (Picea abies) and silver birch (Betula pendula). Tree Physiol 26:605–611
McCarthy S, Ai C, Wheaton G, Tevatia R, Eckrich V, Kelly R, Blum P (2014) Role of an archaeal PitA transporter in the copper and arsenic resistance of Metallosphaera sedula, an extreme thermoacidophile. J Bacteriol Res 196(20):3562–3570
Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014) TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5(1):1–12
Pattanagul W, Thitisaksakul M (2008) Effect of salinity stress on growth and carbohydrate metabolism in three rice (Oryza sativa L.) cultivars differing in salinity tolerance. Indian J Exp Biol 46(10):736–742
Peng P, Gao Y, Li Z, Yu Y, Qin H, Guo Y, Huang R, Wang J (2019) Proteomic analysis of a rice mutant sd58 possessing a novel d1 allele of heterotrimeric G protein alpha subunit (RGA1) in salt stress with a focus on ROS scavenging. Int J Mol Sci 20(1):167
Potashkin JA, Meredith GE (2006) The role of oxidative stress in the dysregulation of gene expression and protein metabolism in neurodegenerative disease. Antioxid Redox Signal 8(1–2):144–151
Rahneshan Z, Nasibi F, Moghadam AA (2018) Effects of salinity stress on some growth, physiological, biochemical parameters and nutrients in two pistachio (Pistacia vera L.) rootstocks. J Plant Interact 13(1):73–82
Ramezani A, Niazi A, Abolimoghadam AA, Babgohari MZ, Deihimi T, Ebrahimi M, Akhtardanesh H, Ebrahimie E (2013) Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress. Mol Biotechnol 53(2):189–197
Shani E, Salehin M, Zhang Y, Sanchez SE, Doherty C, Wang R, Mangado CC, Song L, Tal I, Pisanty O, Ecker JR (2017) Plant stress tolerance requires auxin-sensitive Aux/IAA transcriptional repressors. Curr Biol 27(3):437–444
Shen Y, Du J, Yue L, Zhan X (2016) Proteomic analysis of plasma membrane proteins in wheat roots exposed to phenanthrene. Environ Sci Pollut Res 23(11):10863–10871
Shirokova Y, Forkutsa I, Sharafutdinova N (2000) Use of electrical conductivity instead of soluble salts for soil salinity monitoring in Central Asia. Irrig Drain 14(3):199–206
Singh D, Yadav R, Kaushik S, Wadhwa N, Kapoor S, Kapoor M (2020) Transcriptome analysis of ppdnmt2 and identification of superoxide dismutase as a novel interactor of DNMT2 in the moss Physcomitrella patens. Front Plant Sci 11:1185
Srivastava P, Wu QS, Giri B (2019) Salinity: an overview. In: Giri B, Varma A (eds) Microorganisms in saline environments: strategies and functions. Soil Biology, vol 56. Springer, Cham, Switzerland
Temel A, Janack B, Humbeck K (2017) Drought stress-related physiological changes and histone modifications in barley primary leaves at HSP17 gene. Agronomy 7(2):43
Vaidyanathan H, Sivakumar P, Chakrabarty R, Thomas G (2003) Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)-differential response in salt-tolerant andsensitive varieties. Plant Sci 165:1411–1418
Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93(5):863–873
Vogt JH, Schippers JH (2015) Setting the PAS, the role of circadian PAS domain proteins during environmental adaptation in plants. Front Plant Sci 9(6):513
Wang P, Sun X, Yue Z, Liang D, Wang N, Ma F (2014) Isolation and characterization of MdATG18a, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci Hortic 165:51–61
Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280(4):979–993
Xiong J, Sun Y, Yang Q, Tian H, Zhang H, Liu Y, Chen M (2017) Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci 15(1):19
Zhang Z, Liu Q, Yang J, Yao H, Fan R, Cao C, Liu C, Zhang S, Lei X, Xu S (2020) The proteomic profiling of multiple tissue damage in chickens for a selenium deficiency biomarker discovery. Food Funct 11(2):1312–1321
Acknowledgements
The authors highly acknowledge the Indian Institute of Wheat and Barley Research (IIWBR), Karnal and Central Soil Salinity Research Institute (CSSRI), Karnal, Haryana, India, for providing wheat germplasm for the present study. In addition, the financial assistance to RY in the form of a University Research Scholarship by Maharshi Dayanand University (MDU) is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yadav, R., Santal, A.R. & Singh, N.P. Comparative root proteome analysis of two contrasting wheat genotypes Kharchia-65 (highly salt-tolerant) and PBW-373 (salt-sensitive) for salinity tolerance using LC–MS/MS approach. Vegetos 35, 133–139 (2022). https://doi.org/10.1007/s42535-021-00292-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42535-021-00292-0