Abstract
This article introduces Bayesian inference on the bimodality of the generalized von Mises (GvM) distribution for planar directions (Gatto and Jammalamadaka in Stat Methodol 4(3):341–353, 2007). The GvM distribution is a flexible model that can be axial symmetric or asymmetric, unimodal or bimodal. Two inferential approaches are analysed. The first is the test of null hypothesis of bimodality and Bayes factors are obtained. The second approach provides a two-dimensional highest posterior density (HPD) credible set for two parameters relevant to bimodality. Based on the identification of the two-dimensional parametric region associated with bimodality, the inclusion of the HPD credible set in that region allows us to infer on the bimodality of the underlying GvM distribution. A particular implementation of the Metropolis–Hastings algorithm allows for the computation of the Bayes factors and the HPD credible sets. A Monte Carlo study reveals that, whenever the samples are generated under a bimodal GvM, the Bayes factors and the HPD credible sets do clearly confirm the underlying bimodality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Measurements of directional type appear in many scientific fields: the direction flight of a bird and the direction of earth’s magnetic pole are two examples. These directions can be in the plane, viz. in two dimensions, as in the first example, or they can be in the space, viz. in three dimensions, as in the second example. These measurements are called directional data, and they appear in various scientific fields: in the analysis of protein structure, cf. e.g. Kim et al [23], in machine learning, cf. e.g. Navarro et al [30], in ecology, cf. e.g. Jammalamadaka and Ulric [35], in ornithology, cf. e.g. Schmidt-Koenig [38], in oceanography, cf. e.g. Lin and Dong [26], in meteorology, c.f. e.g. Zhang et al [41], etc. A two-dimensional direction is a point in \({\mathbb {R}}^2\) without magnitude, e.g. a unit vector. It can also be represented as a point on the circumference of the unit circle or as an angle, measured for example in radians and after fixing the null direction and the sense of rotation (clockwise or counterclockwise). Because of this circular representation, observations on two-dimensional directional data are distinctively called circular data. During the last two or three decades, there has been a considerable rise of interest for statistical methods for the analysis of directional data. Recent applications can be found, for example, in Ley and Verdebout [25]. Some monographs on this topic are Mardia and Jupp [27], Jammalamadaka and SenGupta [20], Ley and Verdebout [24] and also Pewsey et al. [32]. A basic introduction is given in Gatto and Jammalamadaka [14], and a recent review can be found in Pewsey and García-Portugués [31]. We refer to Fisher [8] for the presentation of circular data from various scientific fields.
This article considers the problem of Bayesian inference on the bimodality of the generalized von Mises (GvM) circular distribution, which is defined in (1). The number of modes for a circular distribution has practical relevance in various fields. It is the case, for example, in meteorology, precisely in the context of the analysis of directions of winds, and in biology, precisely in the study of directions taken by various animals.
The GvM distribution (of order 2) [13] has density
where \(\mu _1\in [0,2\pi )\), \(\mu _2\in [0,\pi )\) and \(\kappa _1,\kappa _2>0\). The function
is the normalizing constant, where \(\delta =(\mu _1-\mu _2)\text {mod}~\pi \in [0,\pi )\). We denote any circular random variable with this distribution by \(\text {GvM}(\mu _1,\mu _2,\kappa _1,\kappa _2)\).
The GvM is an important extension of the well-known circular normal or von Mises (vM) distribution, which has density
where \(\mu \in [0, 2 \pi )\) is the mean direction, \(\kappa >0\) is the concentration and where \(I_{r}(z)= (2 \pi )^{-1} \int _0^{2 \pi } \cos r \theta \, \exp \{ z \cos \theta \} \mathrm{d} \theta \), \(z \in {\mathbb {C}}\), is the modified Bessel function I of integer order r [1, p. 376]. Thus, \(G_0(\delta ,\kappa ,0)=I_0(\kappa ),~\forall \delta \in [0,\pi ), \kappa >0\). In this case, we denote any circular random variable with this distribution by \(\text {vM}(\mu ,\kappa )\).
Besides its greater flexibility in terms of asymmetry and bimodality, the GvM distribution possesses the following properties that other asymmetric or multimodal circular distributions usually do not have. First, after a re-parametrization, the GvM distribution belongs to the canonical exponential class. In this form, it admits a minimal sufficient and complete statistic. One can refer to Section 2.1 of Gatto and Jammalamadaka [13] for these facts. Second, the maximum likelihood estimator and the trigonometric method of moments estimator of the parameters are the same; cf. Section 2.1 of Gatto [10]. As explained in the next paragraph, the computation of the maximum likelihood estimator is simpler with the GvM distribution than with the mixture of two vM distributions. Third, it is shown in Section 2.2 of Gatto and Jammalamadaka [13] that for fixed trigonometric moments of orders one and two, the GvM distribution is the one with largest entropy. The “maximum entropy principle” for selecting a distribution on the basis of partial knowledge tells that one should always choose distributions having maximal entropy, within distributions satisfying the partial knowledge. In Bayesian statistics, whenever a prior distribution has to be selected and information on the first two trigonometric moments is available, then the GvM is the optimal prior, cf. Berger [4], Section 3.4. For other information theoretic properties of the GvM, see Gatto [11].
Mixture of circular distributions has been proposed in the literature. For example, McVinish and Mengersen [28] examined the use of Bayesian mixtures of triangular distributions in the context of semiparametric analysis of circular data. The mixture of two vM or other distributions provides an alternative bimodal or asymmetric model to the GvM. However, that mixture model does not share the given properties of the GvM. The mixture is not necessarily more practical. While the likelihood of the GvM distribution is bounded, the likelihood of the mixture of the vM(\(\mu _1,\kappa _1\)) and the vM(\(\mu _2,\kappa _2\)) distributions is unbounded. As \(\kappa _1 \rightarrow \infty \), the likelihood when \(\mu _1\) is equal to any one of the sample values tends to infinity. This follows from \(I_0(\kappa _1) \sim ( 2 \pi \kappa _1)^{-1/2} \mathrm{e}^{\kappa _1}\), as \(\kappa _1 \rightarrow \infty \); cf. Abramowitz and Stegun [1], 9.7.1 at p. 377. For alternative estimators to the maximum likelihood for vM mixtures, refer to Spurr and Koutbeiy [39].
The GvM distribution has been applied in various scientific fields, and some recent applications are: Zhang et al. [41], in meteorology, Lin and Dong [26], in oceanography, Astfalck et al. [2], in offshore engineering, and Christmas [7], in signal processing. In spectral analysis of stationary processes, the bimodality of the spectral distribution is an important property. For instance, the frequency domain approach to fatigue life analysis is common in the design of mechanical components that are subject to random loads. In this context, bimodal spectral densities are often observed in the chassis components of road vehicles, cf. Fu and Cebon [9]. In the context of time series, Gatto [12] proposed the GvM spectral distribution as the one that maximizes the spectral entropy. However, the Bayesian literature on the GvM model is practically inexistent: one can only find the Bayesian test on the symmetry of the GvM distribution by Salvador and Gatto [36].
As mentioned, the aim of this article is to introduce Bayesian inference on the bimodality of the GvM distribution. Two approaches of Bayesian inference are considered. The first concerns test of hypotheses, where the null hypothesis \({\hbox {H}}_0\) is the bimodality of (1) and the alternative hypothesis \({\hbox {H}}_1\) is unimodality. This test was motivated by Basu and Jammalamadaka [3], who proposed a similar test for mixtures of vM distributions. The Bayes factor of this test is obtained. The second approach concerns HPD credible set. Two parameters relevant for bimodality are obtained and the HPD credible set for those parameters is compared with the subset of all parameters yielding bimodality.
We can summarize the Bayesian test as follows. Define by \(\varvec{\Psi }\) the set of all vectors of parameters \(\varvec{\psi } = (\kappa _1,\kappa _2,\mu _1,\mu _2)\) of (1) that lead to bimodality and denote the prior probability of bimodality by \(p= P\left[ \varvec{\psi } \in \varvec{\Psi } \right] \). Because we assume that the elements of \(\varvec{\psi }\) are independent, the computation of p can be done by simple Monte Carlo. Assume that the components of the sample \(\varvec{\theta }=(\theta _1,\ldots ,\theta _n)\) are independent and identically distributed (i.i.d.), with GvM distribution (1). Let \(P \left[ B | \varvec{\theta } \right] \) be the conditional probability given \(\varvec{\theta }\), viz. the sample, for any measurable set B. The Bayesian test requires the posterior probability of bimodality \(P \left[ \varvec{\psi } \in \varvec{\Psi } | \varvec{\theta } \right] \).
In the posterior distribution, the components of \(\varvec{\psi }\) are generally not independent. The computation of the posterior probability is done by Markov chain Monte Carlo (MCMC). We remind that MCMC is a class of iterative simulation techniques that is based on three main algorithms: the Metropolis, due to Metropolis et al. [29], the Metropolis–Hastings, due to Hastings [17] and the Gibbs sampler due to Geman and Geman [16]. MCMC algorithms allow to generate a Markov chain whose equilibrium distribution coincides with the posterior distribution. The states of the Markov chain are recorded only after a certain initial period, usually called burn-in period: after this period the generated values of the Markov chain become very close to generations from the posterior distribution.
Precisely, we propose the following nested simulation scheme for the posterior distribution of \(\varvec{\psi }\). The main algorithm is the Gibbs sampler: we generate from the full conditional distributions of each parameter. The full conditional of one element of \(\varvec{\psi }\) is the posterior conditional distribution of that element given all other parameters fixed. The generation from each one of the full conditionals is done with the Metropolis–Hastings (MH) algorithm. Here, the sampling or instrumental density is a piecewise function that interpolates the full conditional.
The simulations from prior and posterior distributions are used to compute prior and posterior probability of bimodality. These probabilities allow us to infer on the bimodality by means of Bayes factors and HPD credible sets.
The next sections of the article present the following topics. Section 2 presents two Bayesian approaches for the bimodality of the GvM distribution: the testing approach, with the Bayes factor, and the HPD approach. It shows how to obtain posterior and full conditional distributions, for some given prior distributions. Then, a nested simulation algorithm from the posterior distribution is explained. It uses the Gibbs sampler and a MH algorithm. Section 3 presents the results of the simulation study. Bayes factors for unimodal and bimodal samples are obtained. HPD credible intervals for single parameters are presented. A bivariate HPD credible set for two parameters that determine bimodality of the GvM is given. Gelman’s convergence diagnostic is presented. This method consists in studying the variance between, the variance within and the shrink factor of m simultaneous chains. We refer to [15] for this. Simulation results under a more general prior are given at the end of the numerical study. Final remarks are given in Sect. 4.
2 Bayesian Inference and MCMC
This section presents the methodology for Bayes inference on bimodality for the GvM, precisely the derivation of BF and HPD credible sets and the related MCMC. Section 2.1 provides the derivation of the posterior distribution. Section 2.2 concerns simulation from the posterior distribution. Its multidimensional and complicated form requires MCMC. We suggest using the Gibbs sampling from the posterior distribution, in conjunction with the MH algorithm for sampling from the one-dimensional full conditionals.
Inferential aspects related to BF and HPD credible sets are explained in Sect. 2.3.
2.1 Likelihood, Posterior Computations and Bimodality
This section provides likelihood of the sample \(\varvec{\theta }=(\theta _1,\ldots ,\theta _n)\) of independent angles from the GvM(\(\mu _1,\mu _2,\kappa _1,\kappa _2\)) distribution and the posterior distribution of the parameters, for some given prior distributions.
The prior distribution of \(\kappa _j\) is the uniform distribution over \(\left( 0,{\bar{\kappa }}_j\right) \), for some \({\bar{\kappa }}_j>0\), for \(j=1,2\). Note that although the major part of the simulation study that follows uses these uniform priors, the more general beta prior is also considered at the end of the simulation study (in Sect. 3.3). The prior distribution of \(\mu _1\) is the \(\text {vM}(\nu _1,\tau _1)\) distribution with density given by (3). Thus,
where \(\tau _1>0\) and \(\nu _1\in [0,2\pi )\).
The prior distribution of \(\mu _2\) is the axial vM with density
where \(\tau _2>0\) and \(\nu _2\in [0,\pi )\) and \(\tau _2>0\). We use the notation \(\mu _2\sim \text {vM}_2(\nu _2,\tau _2)\).
The likelihood of the sample \(\varvec{\theta }=(\theta _1,\ldots ,\theta _n)\) is given by:
The priors (4) and (5) and the likelihood (6), yield the joint posterior density of \((\kappa _1,\kappa _2,\mu _1,\mu _2)\) given by
where I denotes the indicator function. We note in particular that, although \((\kappa _1,\kappa _2,\mu _1,\mu _2)\) are a priori independent, they are a posteriori dependent.
The number of modes of the \(\text {GvM}(\mu _1,\mu _2,\kappa _1,\kappa _2)\) distribution is determined by the nature of the roots of the quartic
where \(b_3=-4\rho \sin \delta \cos \delta \), \(b_2=\rho ^2-1\), \(b_1=2\rho \sin \delta \cos \delta \) and \(b_0=\sin ^2\delta \cos ^2\delta \), with \(\rho =\kappa _1/(4\kappa _2)\) and \(\delta =(\mu _1-\mu _2)\mathrm{mod}\pi \), cf. Gatto and Jammalamadaka [13]. An algebraic analysis that can be found in Salvador and Gatto [37] shows that the null hypothesis of bimodality admits the analytical expression
where
Note that repeated roots are admissible. Unfortunately, the boundary of \({\mathcal {W}}\) does not admit a closed-form expression. We can, however, determine it numerically, and a graphical representation of \({\mathcal {W}}\) is given in Fig. 7a. The alternative hypothesis \({\hbox {H}}_1\) is the general one, namely that the roots are of any other possible nature.
2.2 Computational Aspects
Bayesian inference on the bimodality of the GvM distribution requires simulation from prior and posterior distributions. Prior simulations are straightforward and posterior simulations are done by MCMC.
Besides Monte Carlo, we mention that we compute the normalizing constant of the GvM distribution by
where \(\delta \in [0, \pi )\) and \(\kappa _1,\kappa _2 > 0\), see, e.g. Gatto and Jammalamadaka [13].
2.2.1 Prior Simulation
It follows from the assumed a priori independence of the parameters; their simulation is elementary. For the sake of completeness, we describe here their generation. Simulating the prior probability of bimodality is accomplished with the following simple Monte Carlo algorithm. Let \(\varvec{\psi }=\left( \kappa _1,\kappa _2,\mu _1,\mu _2\right) \).
Computation of prior probability of bimodality by simulation | |
1. Generate \(\varvec{\psi }^{(t)}\), for \(t=1,\ldots ,T\), where each element is generated independently from its | |
one-dimensional prior distribution. | |
2. Compute the prior probability of bimodality by | |
\( \frac{1}{T}\sum _{t=1}^T \mathrm{I}\{ \varvec{\psi }^{(t)} \in {\mathcal {W}}\}. \) |
Simulating \(\kappa _1\) and \(\kappa _2\) is straightforward, as they are assumed uniformly distributed. The simulation of \(\mu _1\) can be done with the program rvonmises of the package Directional in R. Concerning the simulation of \(\mu _2\), from the axial vM distribution, we note that \( \theta \sim \text {vM}(2\nu _2,\tau _2)\Rightarrow \theta /2\sim \text {vM}_2(\nu _2,\tau _2). \) Thus, generations from \(\text {vM}_2(\nu _2,\tau _2)\) are the generations from \(\text {vM}(2\nu _2,\tau _2)\) (with rvonmises) that are then divided by 2.
2.2.2 Posterior Simulation via MCMC
The computation of the posterior probability of bimodality is more complicated. We compute the posterior distribution \(\varvec{\psi }\) by MCMC and precisely by Gibbs sampling (cf. Geman and Geman [16]). By iterative generations from the full conditionals, we obtain a Markov chain whose stationary distribution is the posterior (7) (cf. e.g. Robert and Casella [34], p. 371-424).
The full conditional of \(\kappa _1\), namely the conditional density of \(\kappa _1\) given \(\mu _1,\mu _2, \kappa _2\) and \(\varvec{\theta }\), is given by
The full conditional of \(\kappa _2\) is given by
The full conditional of \(\mu _1\) is given by
The last full conditional is for \(\mu _2\) and it is given by
Given the above full conditionals, the Gibbs sampling scheme is as follows.
Posterior Gibbs sampling of \(\varvec{\psi }\) | |
1. Select an arbitrary starting value \(\varvec{\psi }^{(0)}=\left( \kappa _1^{(0)},\kappa _2^{(0)},\mu _1^{(0)},\mu _2^{(0)}\right) \). | |
2. For \(t=0,\ldots ,N_s-1\), generate iteratively: | |
\(\bullet \) \(\kappa _1^{(t+1)} \text { from } f_1\left( \kappa _1\bigg |\mu _1^{(t)},\mu _2^{(t)},\kappa _2^{(t)},\varvec{\theta }\right) \), | |
\(\bullet \) \(\kappa _2^{(t+1)} \text { from } f_2\left( \kappa _2\bigg |\mu _1^{(t)},\mu _2^{(t)},\kappa _1^{(t+1)},\varvec{\theta }\right) \), | |
\(\bullet \) \(\mu _1^{(t+1)} \text { from } f_3\left( \mu _1\bigg |\mu _2^{(t)},\kappa _1^{(t+1)},\kappa _2^{(t+1)},\varvec{\theta }\right) \), | |
\(\bullet \) \(\mu _2^{(t+1)} \text { from } f_4\left( \mu _2\bigg |\mu _1^{(t+1)},\kappa _1^{(t+1)},\kappa _2^{(t+1)},\varvec{\theta }\right) \). | |
3. For a selected positive integer \(B<N_s\), called burn-in period length, discard \(\{\varvec{\psi }^{(t)}\}_{t=0,\ldots ,B}\). | |
Re-index the \(T=N_s-B\) retained simulations from 1 to T. |
However, the generation from the full conditionals with one of basic methods for univariate distributions (such as inversion, acceptance-rejection) seems neither simple nor efficient. The complication arises from the part \(1/G_0^n(\delta ,\kappa _1,\kappa _2)\). Either by the expansion (10) or by numerical integration of (2), its evaluation is relatively complicated in the context of simulation and it tends to take very small values when n is large. We apply the MH algorithm presented in Robert and Casella [34], p. 276. This algorithm requires a density that is close to the full conditional of interest and from which the simulation is simple. We call this surrogate density as instrumental density, and we present it just after the following MH algorithm.
Univariate MH simulation from full conditionals of \(\varvec{\psi }\) | |
Redenote by \(\psi \) any one of the parameters \(\kappa _1,\kappa _2,\mu _1,\mu _2\) and by \(f(\psi )\) its full conditional. | |
1. Select an arbitrary starting value \(\psi ^{(0)}\). | |
2. For \(t=0,\ldots ,N_s-1\): | |
\(\bullet \) generate \({\tilde{\psi }}_t\) from the instrumental density g, | |
\(\bullet \) set | |
\( \psi ^{(t+1)}= {\left\{ \begin{array}{ll} {\tilde{\psi }}_t,~&{}\text {with probability}~\rho \left( \psi ^{(t)},{\tilde{\psi }}_t\right) ,\\ \psi ^{(t)},~&{}\text {with probability}~1-\rho \left( \psi ^{(t)},{\tilde{\psi }}_t\right) , \end{array}\right. } \) | |
where | |
\( \rho (\psi ,{\tilde{\psi }})=\min \left\{ \frac{f({\tilde{\psi }})}{f(\psi )}\frac{g(\psi ) }{g({\tilde{\psi }})},1\right\} . \) | |
3. For a selected burn-in \(B<N_s\), discard \(\{\psi ^{(t)}\}_{t=0,\ldots ,B}\). | |
Re-index the \(T=N_s-B\) retained simulations from 1 to T. |
Thus, our posterior simulation algorithm has two levels of MCMC.
The instrumental density g in the above MH algorithm is the normalized piecewise linear interpolation of the full conditional. When the knots are \((\psi _0,y_0),(\psi _1,y_1),\ldots ,(\psi _k,y_k)\), it is given by
where \(d_i=(y_{i+1}-y_i)/(\psi _{i+1}-\psi _{i}),~\text {for}~i=0,\ldots ,k-1\).
Let us explain the simulation from the instrumental density of \(\kappa _1\), for example. The interpolation of the full conditional \(f_1(\kappa _1|\mu _1,\mu _2,\kappa _2,\varvec{\theta })\) is taken with 5 interpolation points, A–E in Fig. 1, which are chosen as follows. We fix the first and last points with abscissae values \(\varepsilon =0.2\) and \({\bar{\kappa }}_1\), respectively. The remaining points are generated uniformly in the interval \((\varepsilon , {\bar{\kappa }}_1)\). The instrumental density g is the normalized interpolation function. We generate from g with the inverse transform method. Figure 1 shows the full conditional \(f_1(\kappa _1|\mu _1,\mu _2,\kappa _2,\varvec{\theta })\) and its piecewise interpolation function.
2.3 Bayes Factor, HPrD and HPD Credible Sets
This section briefly summarizes the definitions of the Bayes factor, of the HPrD and of the HPD credible sets.
In our setting, the Bayes factor \({\hbox {B}}_{01}\) is an indicator of the support of the sample for the null hypothesis \({\hbox {H}}_0\) of bimodality of the GvM, against the alternative hypothesis \({\hbox {H}}_1\) of unimodality. It is precisely the ratio of the posterior odds over the prior odds, of \({\hbox {H}}_0\) versus \({\hbox {H}}_1\), which is symbolically given by
This can obviously be re-expressed as
When \({\hbox {B}}_{01}>1\), the sample supports \({\hbox {H}}_0\). When \({\hbox {B}}_{01}<1\), the sample supports \({\hbox {H}}_1\). Table 1 gives some practical guidelines for the values Bayes factors; we refer also to Kass and Raftery [22] and Jeffreys [21].
Alternatively to the Bayes factor, Bayesian inference can rely on the HPD credible set. We firstly introduce a general concept. Let f be a density over \({\mathbb {R}}\). The highest density region (HDR) at level \(1-\alpha \) is the set
where \(f_{\alpha }\) is the largest constant such that \(P\left[ R\left( f_{\alpha }\right) \right] \ge 1-\alpha \), for some (typically small) \(\alpha \in (0,1)\); cf. e.g. Hyndman [19]. In particular, it follows that, amongst all regions with coverage probability \(1-\alpha \), the HDR has the smallest volume. Computational aspects of HDR are studied in Wright [40], Hyndman [18] and Hyndman [19].
In the Bayesian setting where f is the posterior density, the HDR is called HPD credible set; cf. e.g. Box and Tiao [5]. When f is the prior density, we call the HDR as the highest prior density (HPrD) credible set. We refer to Chen and Shao [6] for computational aspects of HPD credible sets. Bimodality of the GvM is related to the parameters \(\rho \) and \(\delta \), as explained in Sect. 2.1. Thus, Bayesian inference on bimodality can rely on bivariate HPD credible sets for \((\rho ,\delta )\).
3 Simulation Study
In this section, we apply the methods that we proposed for the inference on the bimodality of the GvM: the MCMC algorithm of Sect. 2.2.2, the convergence diagnostics by Gelman et al. [15] and the Bayes factors and HPD of Sect. 2.3.
We use various simulated samples \(\varvec{\theta }= (\theta _1,\ldots ,\theta _n)\). The sample values are generated independently from the following distributions.
-
A sample \(\varvec{\theta }\) of \(n=100\) values is generated from the unimodal \(\text {GvM}(\pi ,\pi /2,1.2,0.2)\) in Sect. 3.1.1, for the Bayes factor. This sample is also used in Sect. 3.3.1, for the Bayes factor with an alternative prior.
-
A sample \(\varvec{\theta }\) of \(n=100\) values is generated from the bimodal \(\text {GvM}(\pi ,\pi /2,0.16,0.2)\) in Sect. 3.1.2, for the Bayes factor. This sample is also used in Sect. 3.3.3, for the Bayes factor with an alternative prior.
-
A sample \(\varvec{\theta }\) of \(n=100\) values is generated from the bimodal \(\text {GvM}(\pi ,\pi /2,1.5,1.5)\) in Sect. 3.2, for the HPD credible set, and also used at the end of Sect. 3.2, for convergence diagnostic.
-
Two samples \(\varvec{\theta }\) of \(n=10\) and \(n= 200\) values are additionally generated from the same bimodal \(\text {GvM}(\pi ,\pi /2,1.5,1.5)\) in Sect. 3.2, for the study of the influence of the sample size n on the HPD credible intervals.
-
A sample \(\varvec{\theta }\) of \(n=200\) values is generated from the unimodal \(\text {GvM}(\pi ,\pi /2,1.2,0.2)\) in Sect. 3.3.2, for the Bayes factor with an alternative prior.
All these generated samples are displayed through their kernel density estimations.
The hyperparameters for the prior specifications are chosen as follows:
Prior and posterior probabilities of bimodality are obtained by simple Monte Carlo and Gibbs sampling, respectively, as explained in Sect. 2.2. The number of simulations is \(T=4\times 10^4\) for the prior, and for the posterior we have \(N_s=5\times 10^4\) minus \(B=10^4\) retained simulations, namely total simulation minus the burn-in simulations. We choose \(\varvec{\psi }^{(0)}=(\kappa _1^{(0)},\kappa _2^{(0)},\mu _1^{(0)}, \mu _2^{(0)})=(1.5,1.5,\pi ,\pi /2)\) as starting point for all Markov chains. Our Bayes factors and HPD credible sets are based on one single Markov chain. However, for the purpose of Gelman’s convergence diagnostic, \(m=5\) Markov chains are generated with the same starting point \(\varvec{\psi }^{(0)}\).
This computational intensive Monte Carlo study was run with the software R on a main computer cluster. Computer programs are available at http://www.stat.unibe.ch.
3.1 Bayes Factor
This section presents simulation results for the Bayes factor of the test of bimodality of the GvM. For the given sample \(\varvec{\theta }\) from a GvM, we compute prior and posterior probabilities of bimodality by simulating \(\{\varvec{\psi }^{(t)}\}_{t=1,\ldots ,T}\) from the prior and posterior distributions, respectively, with the algorithms of Sect. 2.2. These prior and posterior simulations are iterated \(N=500\) times, and the retained Bayes factor is the average of N simulated Bayes factors. The section is divided into two parts: in part 3.1.1 we generate the sample \(\varvec{\theta }\) from the unimodal GvM, whereas in part 3.1.2 we generate the sample \(\varvec{\theta }\) from the bimodal GvM.
3.1.1 Unimodal GvM Sample
We generate the sample \(\varvec{\theta }\) of size \(n=100\) from the unimodal \(\text {GvM}(\pi ,\pi /2,1.2,0.2)\). Unimodality is formally justified by the fact that the point \((\rho ,\delta )=(1.5,\pi /2)\) is really in the region \({\mathcal {W}}^{{\mathsf {c}}}\) of unimodality. This region is shown in white in Fig. 7a only for values of \(\rho \) smaller than 1.25, but for values larger than 1.25 the region does remain white. The kernel density estimation of this sample is given in Fig. 2a, which confirms unimodality.
We compute the representative Bayes factor \({\bar{{\hbox {B}}}}_{01}\) by the mean of the generated Bayes factors \(B_i,~i=1,\ldots ,N\). The \(N=500\) prior probabilities of bimodality are obtained by simple Monte Carlo; cf. Sect. 2.2.1. The average of these N values is \({\bar{P}}\left[ \text {H}_0\right] =0.802\): there is a priori support for the null hypothesis of bimodality.
The posterior probability of bimodality \({\bar{P}}\left[ \text {H}_0|\varvec{\theta }\right] =0.150\) is also obtained by averaging N values that are simulated by the MCMC algorithm of Sect. 2.2.2. The Bayes factor obtained with this study is given by
The aim of the process of averaging \(N=500\) values is the control of the simulation variability. The boxplot of the generated N values of \({\hbox {B}}_{01}\) can be found in Fig. 2b. The asymptotic normal confidence interval at level 0.95 for the Bayes factor is given by \( \text {I}= {\bar{{\hbox {B}}}}_{01}\pm 1.96 N^{-1/2} \left\{ N^{-1}\sum _{i=1}^{N}({\bar{{\hbox {B}}}}_{01}-B_i)^2 \right\} ^{1/2}\). Obviously, although the N Bayes factors may not be normally distributed, their mean is asymptotically normal. We thus obtain the confidence interval
As the sample \(\varvec{\theta }\) is generated from a unimodal GvM, our prior evidence of bimodality is no longer supported by the sample and thus \({\bar{{\hbox {B}}}}_{01}\) does not support bimodality. The Bayes factor gives negative evidence of bimodality according to Table 1. Note that very few large values are discarded from Fig. 2b, but they are nevertheless considered in the computation of the average Bayes factor and of the confidence interval.
3.1.2 Bimodal GvM Sample
We now consider a sample of size \(n=100\) \(\varvec{\theta }\) generated from a bimodal GvM, precisely from \(\text {GvM}(\pi ,\pi /2,0.16,0.2)\). We compute \((\rho ,\delta )=(0.2,\pi /2)\), which belongs to the region of bimodality \({\mathcal {W}}\) shown in grey in Fig. 7a. The kernel density estimation of \(\varvec{\theta }\) shown in Fig. 3a is also indicating bimodality. As before, we compute the representative Bayes factor by the mean of \(N=500\) values. The prior probability is the same as in 3.1.1, and it is \({\bar{P}}\left[ \text {H}_0\right] =0.802\). The posterior probability of bimodality \({\bar{P}}[\text {H}_0 |\varvec{\theta }]=0.992\) and thus we obtain
As in Sect. 3.1.1, we compute the asymptotic normal confidence interval for the Bayes factor at level 0.95. It is given by
In this case, the Bayes factor gives decisive evidence of bimodality. Figure 3b shows the boxplot of the Bayes factors. Here also, very few large values are discarded from the boxplot but are considered for the average of the Bayes factors and for the confidence interval.
3.1.3 Remark: Non-GvM Sample
In this remark, we study the behaviour of this test when the sample does not follow the assumed GvM distribution and thus when the given likelihood is inappropriate. We generate a sample of size \(n=100\) from the mixture of three vM distributions given by
with
The density estimation of this non-GvM sample is given in Fig. 4a, and we can observe some clear evidence of trimodality. Figure 4b shows the boxplot of \(N=200\) simulated Bayes factors. Their mean value is
This value lies just within the range of positive evidence of bimodality of Table 1. Thus, two of the three modes have been associated with a single one, but it appears difficult to tell which ones they are. Perhaps the two modes of similar height (at \(\eta _1\) and \(\eta _2\)) have been confounded or perhaps the lower mode (at \(\eta _3\)) has been neglected. Although we cannot provide an unambiguous explanation, we note that the amount of positive evidence for bimodality is minor. The behaviour of the test in this erroneous situation seems reasonable.
This test of bimodality is indeed constructed around the GvM distribution. The null hypothesis is characterized by the region of bimodality of the GvM, \({{{\mathcal {W}}}}\) given in (9), which is obtained from the nature of the roots of the quartic (8). The adequacy of the sample with the GvM model could be checked with the goodness-of-fit tests of Section 7.2 of Jammalamadaka and SenGupta [20]. Let us remind that various types of tests are available: Kuiper’s test (the circular version of Kolmogorov–Smirnov test), Watson’s test (the circular version of Cramer–von Mises test), Ajne’s test (the circular version of Chi-square test) and Rao’s test (based on spacings between sample angles). The GvM distribution function is required for some of these tests, and it can be found in Section 3.3 of Gatto [12].
3.2 HPD and HPrD Credible Sets
This section provides a simulation of the HPD credible set of \((\rho ,\delta )\), which allows to determine the a posteriori evidence of uni- or bimodality. It also provides the simulation of HPrD and HPD credible intervals for \(\kappa _1,\kappa _2,\mu _1, \mu _2\). One sample \(\varvec{\theta }\) of size \(n=100\) is considered, and it is generated under the bimodal \(\text {GvM}(\pi ,\pi /2,1.5,1.5)\). Its kernel density estimation is shown in Fig. 5b, and we can see that bimodality is preserved.
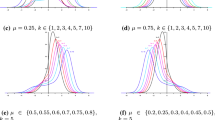
We begin with the presentation of the HPrD and HPD credible intervals. These intervals are given in Table 2. We see that all HPD credible intervals are narrower than the HPrD credible intervals and that they contain their true value. The level is 0.95. Then, we illustrate the effect of the sample size n on the HPD credible interval by considering two additional samples of sizes \(n=10\) and 200 that are generated from the same \(\text {GvM}(\pi ,\pi /2,1.5,1.5)\). Their density estimation is given in Fig. 5a, c, respectively, and we see that bimodality is preserved. We restrict the analysis to the parameter \(\mu _1\). The results given in Table 3 show that, as n increases, the HPD credible intervals at level 0.95 become narrower around the true value. A graph of this can be found in Fig. 6, where the posterior densities, HPrD and HPD credible intervals are given. The grey regions have area 0.95, and they are based over the HPD credible intervals. The HPrD credible intervals are displayed with a bold line along the abscissae. As the sample size n increases, the HPD credible intervals become narrower around the true value of the parameter. Computations are done using emp.hpd of the R-package TeachingDemos, with \(\alpha =0.05\).
As mentioned in Sect. 2.1, the bimodality of the GvM is determined by the number of real roots in \([-1,1]\) of the quartic \(q_{\rho ,\delta }\) in (8). We can infer on the bimodality of the GvM through the bivariate HPD credible set of \((\rho ,\delta )\). We use the same sample \(\varvec{\theta }\) of size \(n=100\). The level of the HPD credible set is 0.95, and we compute the HPD with the function HPDregionplot of the R-package emdbook. Figure 7a shows that the HPD credible set obtained from the simulation is completely included in \({\mathcal {W}}\), the region of bimodality of the set of the parameters \((\rho ,\delta )\) given in (9). This region of bimodality is shown in grey. The graph with the region of bimodality is due to Salvador and Gatto [37] and Pfyffer and Gatto [33]. Thus, the HPD credible set gives us a clear confirmation of the bimodality. Figure 7b shows the posterior of \((\rho ,\delta )\) and indicates that the HPD of Fig. 7a should maintain the connected form at different levels.
We now verify the convergence of the simulations. We consider only the sample \(\varvec{\theta }\) of size \(n=100\), which is also used in the first part of this section.
In order to apply Gelman’s diagnostic, we generate \(m=5\) chains. According to Gelman et al. [15], the values of the variance between b, the variance within w and the shrink factor r in Table 4 confirm convergence for each one of the four components of the chain. Thus, convergence of the whole Markov chain \(\{\varvec{\psi }^{(t)}\}_{t=1,\ldots ,T}\) is achieved. A visual representation of the convergence is done using the command gelman.plot of the package coda that plots the shrink factor r evaluated at different instants of the simulation. The Markov chains are divided into bins according to the parameters bin.width and max.bins, which are the number of simulations per segment and the maximum number of bins, respectively. The shrink factor is repeatedly calculated. The first shrink factor is calculated with 50 simulations, the second with \(50+\texttt {bin.width}\) simulations, the third with \(50+2\cdot \texttt {bin.width}\) simulations, and so on. In Fig. 8, we can see how the shrink factor of each parameter \(\kappa _1\), \(\kappa _2\), \(\mu _1\), \(\mu _2\) evolves with respect to the iterations of the chain: the convergence of the chains associated with \(\kappa _1\) and \(\kappa _2\) appears slower, since at the beginning the shrink factors fluctuate. But then they stabilize around the value 1. Thus, we can consider the test passed and convergence is achieved.
Visuals results for the convergence of the single parameters can be found in Fig. 9. The four histograms of the simulations are close to four graphs of the full conditionals (solid line), indicating that each one of the four components of the Markov chain converges to the respective full conditional.
3.3 More General Prior
So far, the prior distribution of \(\kappa _j\) has been the uniform distribution over \(\left( 0,{\bar{\kappa }}_j\right) \), for some \({\bar{\kappa }}_j>0\), for \(j=1,2\). This choice may not always be convenient mainly because of the abrupt jump of the densities at \({\bar{\kappa }}_1\) and \({\bar{\kappa }}_2\), making the choice of these values rather consequent. Because we may want a prior that decays slowly on the right tail, we can consider the two beta priors with densities proportional to \(\left( \kappa _j/{\bar{\kappa }}_j\right) ^{\alpha _j -1} \left( 1-\kappa _j/{\bar{\kappa }}_j\right) ^{\beta _j-1}\), \(\forall \kappa _j \in [0, {\bar{\kappa }}_j]\), with \(\alpha _j,\beta _j >0\), for \(j=1,2\). Thus, the uniform priors used so far and given in Sect. 2.2.1 are retrieved with \(\alpha _j=\beta _j=1\), for \(j=1,2\).
In the simulation study, we select \(\alpha _j=\beta _j=2\) and, as before, \({\bar{\kappa }}_j=3\), for \(j=1,2\). The priors for \(\mu _1,\mu _2\) are taken as in Sect. 2.2.2. All hyperparameters are as in Sect. 3.
Thus, the full conditional of \(\kappa _1\) is given by
and the full conditional of \(\kappa _2\) is
The full conditionals of \(\mu _1\) and \(\mu _2\) are as in Sect. 2.2.2.
We use the unimodal sample of size \(n=100\) and the bimodal sample of size \(n=100\) that are used in Sects. 3.1.1 and 3.1.2, respectively. Moreover, we consider a third additional unimodal sample with larger size \(n=200\).
3.3.1 Unimodal Sample of Size \(n=100\)
We first consider the same unimodal sample of size \(n=100\) of Sect. 3.1.1, which is generated from the \(\text {GvM}(\pi ,\pi /2,1.2,0.2)\) distribution. The density estimation with this sample is given in Fig. 2a. Prior and posterior simulations are iterated \(N = 500\) times. These simulations allow us to compute the prior probability of bimodality \({\bar{P}}[\text {H}_0]=0.882\) and the posterior analogue \({\bar{P}}[\text {H}_0|\varvec{\theta }]=0.784\). The mean value of the \(N=500\) generated Bayes factors is
and the asymptotic normal confidence interval at level 0.95 for the true Bayes factor is
The boxplot of the resulting Bayes factors is given in Fig. 10a. The posterior probability of bimodality \({\bar{P}}[{\hbox {H}}_0|\varvec{\theta }]\) is indeed smaller than its prior \({\bar{P}}[{\hbox {H}}_0]\), but not much smaller. The mean Bayes factor shows negative evidence of bimodality, but its value is quite larger than the value obtained in Sect. 3.1.1.
3.3.2 Unimodal Sample of Size \(n=200\)
In order to clarify the question of the size of the previous Bayes factor, we increase the sample size to \(n=200\). A new sample \(\varvec{\theta }\) of size \(n=200\) is generated from \(\text {GvM}(\pi ,\pi /2,1.2,0.2)\). Its kernel density estimation is given in Fig. 10c. The posterior probability is now given by \({\bar{P}}[{\hbox {H}}_0|\varvec{\theta }]=0.0519\). The previous prior probability \({\bar{P}}[{\hbox {H}}_0]=0.882\) is considered. The mean of \(N=500\) simulated Bayes factors is
and the asymptotic normal confidence interval at level 0.95 is
The Bayes factor gives again negative evidence of bimodality, but it is now more substantial. The boxplot of the simulated Bayes factors can be found in Fig. 10c. The Bayes factor and the posterior probability are significantly lower than with the sample of size \(n=100\). This is what we would have expected. The prior probability of bimodality \({\bar{P}}[{\hbox {H}}_0]=0.882\) is bigger than the one of Sect. 3.1.1. Thus, we need an unimodal sample of larger size in order to obtain a sufficiently small posterior probability of bimodality.
3.3.3 Bimodal Sample of Size \(n=100\)
We now consider the bimodal sample used in Sect. 3.1.2, of \(n=100\) replications generated from \(\text {GvM}(\pi ,\pi /2,0.16,0.2)\). The density kernel estimation of the sample can be found in Fig. 3a. The prior probability of bimodality does not change: it is given by \({\bar{P}}[\text {H}_0]=0.882\). The posterior probability of bimodality in this obtained by \(N=500\) simulations is \({\bar{P}}[\text {H}_0|\varvec{\theta }]=0.997\). This leads to the mean of \(N=500\) Bayes factors
and to the asymptotic normal confidence interval at level 0.95
The Bayes factor gives thus decisive evidence of bimodality. The boxplots of the resulting Bayes factors can be found in Fig. 10d.
4 Final Remarks
This article studies Bayesian inference on the bimodality of the GvM distribution: MCMC algorithms for computing Bayes factors and HPD regions are proposed and tested by simulation. Thus, it extends the list of available inferential methods for the GvM model: trigonometric method of moments estimation, cf. Gatto [10], and Bayesian test of symmetry, cf. Salvador and Gatto [36].
In Sect. 1, various interesting properties of the GvM model are mentioned together with the importance of bimodality in various applied fields. For example, data on wind directions Gatto and Jammalamadaka [13] show bimodality. Bimodality is indeed more frequent and important with circular than it is with linear data. In this context, the proposed Bayesian tests are practically relevant.
One may extend this study in two ways. First by proposing a frequentist test of bimodality. One could write a likelihood ratio test and exploit the partition of the parametric set in terms of unimodality and bimodality which is shown in Fig. 7a. A second and perhaps more challenging way of extending this study would be by considering prior distributions with dependent GvM parameters. One would thus generalize the full conditionals and the MCMC algorithm of Sect. 2.2.2. We would then obtain circular–circular, circular–linear or other unusual types of joint distributions. A difficulty would be to obtain specific simulation methods from these joint distributions. Another difficulty would be the quantification of the dependence of the joint distributions on these manifolds. We could use or adapt the circular–circular and circular–linear correlation coefficients that are given, for example, in Sections 8.2–8.5 of Jammalamadaka and SenGupta [20].
References
Abramowitz M, Stegun IA (1965) Handbook of mathematical functions: with formulas, graphs, and mathematical tables, vol 55. Courier Corporation
Astfalck L, Cripps E, Gosling J, Hodkiewicz M, Milne I (2018) Expert elicitation of directional metocean parameters. Ocean Eng 161:268–276
Basu S, Jammalamadaka S (2000) Unimodality in circular data: a Bayes test. Adv Methodol Appl Aspects Probab Stat 1:4–153
Berger JO (1985) Statistical decision theory and Bayesian analysis. Springer
Box GE, Tiao GC (1973) Bayesian inference in statistical analysis. Addision-Wesley, Reading
Chen M-H, Shao Q-M (1999) Monte Carlo estimation of Bayesian credible and HPD intervals. J Comput Graph Stat 8(1):69–92
Christmas J (2014) Bayesian spectral analysis with Student-t noise. IEEE Trans Signal Process 62(11):2871–2878
Fisher NI (1995) Statistical analysis of circular data. Cambridge University Press
Fu T-T, Cebon D (2000) Predicting fatigue lives for bi-modal stress spectral densities. Int J Fatigue 22(1):11–21
Gatto R (2008) Some computational aspects of the generalized von Mises distribution. Stat Comput 18(3):321–331
Gatto R (2009) Information theoretic results for circular distributions. Statistics 43(4):409–421
Gatto R (2022) Chapter. 10 Information theoretic results for stationary time series and the Gaussian-generalized von Mises time series. In Sen Gupta A, Arnold B (eds) Directional statistics for innovative applications. A bicentennial tribute to Florence Nightingale. Springer
Gatto R, Jammalamadaka SR (2007) The generalized von Mises distribution. Stat Methodol 4(3):341–353
Gatto R, Jammalamadaka SR (2014) Directional statistics: introduction. Statistics Reference Online, Wiley StatsRef, pp 1–8
Gelman A, Rubin DB et al (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7(4):457–472
Geman S, Geman D (1984) Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell PAMI 6(6):721–741
Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57(1):97–109
Hyndman RJ (1990) An algorithm for constructing highest density regions. Technical report, Department of Statistics, University of Melbourne
Hyndman RJ (1996) Computing and graphing highest density regions. Am Stat 50(2):120–126
Jammalamadaka SR, SenGupta A (2001) Topics in circular statistics, vol 5. World Scientific
Jeffreys H (1939) Theory of probability. Oxford University Press, London
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90(430):773–795
Kim S, SenGupta A, Arnold BC (2016) A multivariate circular distribution with applications to the protein structure prediction problem. J Multivar Anal 143:374–382
Ley C, Verdebout (2017) Modern directional statistics. CRC Press
Ley C, Verdebout T (2018) Applied directional statistics: modern methods and case studies. CRC Press
Lin Y, Dong S (2019) Wave energy assessment based on trivariate distribution of significant wave height, mean period and direction. Appl Ocean Res 87:47–63
Mardia KV, Jupp PE (2000) Directional statistics, vol 494. Wiley
McVinish R, Mengersen K (2008) Semiparametric Bayesian circular statistics. Comput Stat Data Anal 52(10):4722–4730
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21(6):1087–1092
Navarro AKW, Frellsen J, Turner RE (2017) The multivariate generalised von Mises distribution: inference and applications. In: Proceedings of the thirty-first AAAI conference on artificial intelligence, AAAI’17. AAAI Press, pp 2394–2400
Pewsey A, García-Portugués E (2021) Recent advances in directional statistics. TEST, pp 1–58
Pewsey A, Neuhäuser M, Ruxton GD (2013) Circular statistics in R. Oxford University Press
Pfyffer S, Gatto R (2013) An efficient simulation algorithm for the generalized von Mises distribution of order two. Comput Stat 28(1):255–268
Robert C, Casella G (2013) Monte Carlo statistical methods. Springer
Jammalamadaka SR, Ulric L (2006) The effect of wind direction on Ozone levels- a case study. Environ Ecol Stat 13:287–298
Salvador S, Gatto R (2022) Bayesian tests of symmetry for the generalized von Mises distribution. Comput Stat 37:947–974. https://doi.org/10.1007/s00180-021-01147-7
Salvador S, Gatto R (2021) An algebraic analysis of the bimodality of the generalized von Mises distribution. Institute of mathematical statistics and actuarial science, Universty of Bern, Preprint
Schmidt-Koenig K (1963) On the role of loft, the distance and site of release in pigeon homing (the “cross-loft experiment’’). Biol Bull 125:154–164
Spurr BD, Koutbeiy MA (1991) A comparison of various methods for estimating the parameters in mixtures of von Mises distributions. Commun Stat-Simul Comput 20(2–3):725–741
Wright D (1986) A note on the construction of highest posterior density intervals. J R Stat Soc Ser C (Appl Stat) 35(1):49–53
Zhang L, Li Q, Guo Y, Yang Z, Zhang L (2018) An investigation of wind direction and speed in a featured wind farm using joint probability distribution methods. Sustainability 10(12)
Funding
Open access funding provided by University of Bern.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gatto, R., Salvador, S. Bayesian Inference on the Bimodality of the Generalized von Mises Distribution. J Stat Theory Pract 16, 32 (2022). https://doi.org/10.1007/s42519-022-00250-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s42519-022-00250-2
Keywords
- Bayes factor
- Circular distribution
- Full conditional distribution
- Gibbs sampling
- Highest posterior density credible set
- Metropolis–Hastings algorithm