Abstract
One of the areas with the potential to be explored in quantum computing (QC) is machine learning (ML), giving rise to quantum machine learning (QML). In an era when there is so much data, ML may benefit from either speed, complexity or smaller amounts of storage. In this work, we explore a quantum approach to a machine learning problem. Based on the work of Mari et al., we train a set of hybrid classical-quantum neural networks using transfer learning (TL). Our task was to solve the problem of classifying full-image mammograms into malignant and benign, provided by BCDR. Throughout the course of our work, heatmaps were used to highlight the parts of the mammograms that were being targeted by the networks while evaluating different performance metrics. Our work shows that this method may hold benefits regarding the generalization of complex data; however, further tests are needed. We also show that, depending on the task, some architectures perform better than others. Nonetheless, our results were superior to those reported in the state-of-the-art (accuracy of 84% against 76.9%, respectively). In addition, experiments were conducted in a real quantum device, and results were compared with the classical and simulator.
Similar content being viewed by others
1 Introduction
It was in 1981 when Feynman (1982) first proposed a basic model for a quantum computer, tackling the inability of classical computers to simulate the physical world. In 1985, David Deutsch proposed the quantum Turing machine, thus formalizing a universal quantum computer (Deutsch 1985). The potential of quantum computers was later proven by Shor (1999) in 1994. Shor developed an algorithm that solves in polynomial time prime factorization and discrete logarithms, an exponential advantage over known classical algorithms. Shortly after, in 1996, another major step was taken by Grover (1996), by developing an algorithm that finds a given value in an array in \(\mathcal {O}(\sqrt {N})\) steps, where N is the array size. Classical algorithms will take a minimum of N/2 steps.
Since then, a lot of progress has been made, particularly in the past decade. Nowadays, there is an ongoing investment by technology companies such as Google, Microsoft, IBM, or D-Wave, making it possible to access cloud-based quantum computers. One of the most promising applications of quantum computing is in ML. Depending on the model, computational complexity may be an obstacle. Recent studies, such as the one performed by IBM and MIT (Havlíček et al. 2019) or the one published by Biamonte et al. (2017), conclude that ML could benefit from the exponentially large quantum state space through controllable entanglement and interference, bringing speed and efficiency to the process.
In ML, different types of neural networks models have been broadly explored, such as convolutional neural networks (CNN) or recurrent neural networks (RNN). A good example of such models for QML are CNNs. CNNs are one of the best algorithms in regard to image content identification and have shown exemplary performance in several tasks (Huang et al. 2017). However, one of its known disadvantages is its complexity. The further we go into a neural network, the more complex are the features it can recognize since they end up aggregating and recombining. Improving the speed of these networks can have a huge impact when training models that require detailed images as input, such as mammograms.
Breast cancer is the type of cancer with the most incidence in women worldwide, with about 1.7 million new cases diagnosed in 2012, representing roughly 25 percent of all cancers in women. It is also the second most frequent cause of cancer death in women, after lung cancer (Fund 2020). In this research, we present an approach where we make use of a quantum method in the aid of breast cancer screening. Our proposal is inspired by the work of Mari et al. (2019) on TL in hybrid classical-quantum neural networks. Our task was to classify full-image mammograms using pretrained classical neural networks resourcing to a quantum enhanced TL method. The advantage of the TL method, is that, it can be applied to a task where a model can be reused as the starting point on another task. In contrast to the work of Mari et al. (2019), we evaluate large images, compare our results across different architectures, and, using the top performer, we apply a series of tests to evaluate the model against its classical counterparts, using different performance metrics. We conduct classical experiments without and with TL where we can noticed the advantage of resorting to TL, for example, through accuracy 67% versus 84%, respectively. In addition, we resort to a quantum simulator varying the circuit depth from 1 to 4 where we also observe an accuracy of 84% for depth equals 1 and 4. Finally, we use a real quantum device where we observe an accuracy of 81%. These results are very promising, since we are in the noisy intermediate-scale quantum (NISQ) era (Preskill 2018), where quantum devices are associated with the need for error correction and still are in their early stage. Moreover, the quantum models version showed a faster training comparing to the classical version which is also quite interesting.
The research work is structured as follows: in Section 2 we show the related work in the field, in Section 3 we introduce the details on the QML topic. Section 4 describes the experimental setup. The summarizing of the data used, test of the proposed model, and results are in Section 5. Finally, in Section 6, we outline the resulting contribution and present some future work.
2 Related work
In this section we summarize the research work on imaging on the topics QML, TL from a clinical perspective, and also, breast cancer screening using deep learning (DL). In order to compare the quality of the classifiers, we will be using common metrics to assess classification models. In what follows, we consider binary classification, where true positive (TP) corresponds to the number of positive examples that are predicted as being positive, true negative (TN) corresponds to negative examples that are predicted as negative, false positive (FP) corresponds to a negative example that was predicted as a positive, and false negative (FN) corresponds to a positive example that was predicted as a negative (Krishna et al. 2018). Based on these counters, the following metrics are defined:
-
Sensitivity, recall or true positive rate (TPR): measures the rate of actual positives that are acknowledged as positives by the classifier.
$$ \frac{TP}{TP + FN} $$ -
Specificity or true negative rate (TNR): measures the classifier’s capacity to isolate negative results.
$$ \frac{TN}{TN + FP} $$ -
Precision: measures the rate of correctly classified instances for one class.
$$ \frac{TP}{TP + FP} $$ -
Accuracy: measures the amount of correctly classified instances of any class.
$$ \frac{TP + TN}{TP + TN + FP + FN} $$ -
F1-score: is the harmonic mean of precision and recall, with equal weights for both.
$$ 2*\frac{precision*recall}{precision+recall} $$ -
Area under the curve (AUC): AUC represents the degree of separability between classes. It makes use of receiver operating characteristics (ROC) curve, which measures TPR, in the y-axis, against FPR (1-TPR), in the x-axis. AUC is a very common indicator of quality performance used in image classification.
2.1 Imaging in quantum machine learning
In an initial search on image processing algorithms for circuit-based quantum models, it is clear that there are several interesting papers solving tasks such as the algorithm proposed by Duan et al. (2019) for dimensionality reduction, the quantum feature extraction framework proposed by Zhang et al. (2015), the quantum representation of color digital images presented by Sang et al. (2017) or the quantum image edge extraction algorithm proposed by Gao et al. (2010) based on improved Sobel operator.
In regard to circuit-based quantum neural networks, Henderson et al. (2020) empirically evaluated the potential benefit of quanvolutional layers by comparing three types of models built on the MNIST dataset: CNNs, QNN (quantum neural networks), and CNN with additional non-linearities introduced, concluding that QNN models showed a faster training. Grant et al. (2018) used MNIST, Iris, and a synthetic dataset of quantum states to compare the performance for several different parameterizations. Tang and Shu (2014) used rough sets (RS) and QNN in order to recognize electrocardiogram (ECG) signals. Zhang et al. (2020) proved that when compared to the random structure QNN, QNN with tree tensor (TT-QNN) architectures have gradients that vanish polynomially with the qubit number showing better trainability and accuracy for binary classification.
Skolik et al. (2020) focus on solving the problem of barren plateaus of the error surface caused by the low depth of circuits by incrementally growing circuit depth during optimization and updating subsets of parameters in each training step. Kerenidis et al. (2019) proposed a quantum algorithm for evaluating and training deep convolutional neural networks for both the forward and backward passes, providing practical evidence for its efficiency using the MNIST dataset. Kaur et al. (2018) presented a novel ensemble-based quantum neural network in order to overcome the over-fitting issues present in speaker recognition techniques.
2.2 Transfer learning in clinical imaging
Several studies (Chen et al. 2019; Kim et al. 2017; Shie et al. 2015) support the use of TL using CNN for the analysis of medical images. Certain methods are more frequently employed according to the clinical object of study (e.g., brain, breast, eye), the image acquisition method (e.g., X-rays, ultrasound, or magnetic resonance), the depth of the sample, and the size of the dataset.
Although there is no validated proof of which method works best for a given clinical problem, Morid et al. (2020) suggest that AlexNET is the most commonly CNN model used for brain magnetic resonance images (Lu et al. 2019; Wang et al. 2019) and breast X-rays (Lévy and Jain 2016; Omonigho et al. 2020), while DenseNet for lung X-rays (Liu et al. 2019; Yan et al. 2019) and shallow CNN models for skin and dental X-rays (Nunnari et al. 2020; Zhang et al. 2019). In addition, with smaller datasets, the most frequently applied TL approach is feature extracting, while fine-tuning is more used with larger datasets; additionally, data augmentation (e.g., translation, rotation of images), as a mean to feed more artificially generated samples to the CNN model in exchange of computational stress, is most frequently employed along with fine-tuning TL approach. Furthermore, studies using fine-tuning TL approaches use fully connected layers (contrary to traditional classifiers) more often than studies employing feature extracting TL approaches, which is due to the fact that training fully connected layers usually requires larger datasets when compared to training traditional classifiers.
In brief, the majority of studies do not benchmark their CNN model against any other model, and the few that actually do, compare against only one model (Morid et al. 2020). Quantum transfer learning introduces low-depth quantum circuits as a subroutine for a classical model. In this context, Mari et al. (2019) propose a method that focuses on the paradigm in which a pretrained classical network is modified and amplified by a final variational quantum circuit.
Moreover, Acar and Yilmaz (2020) applied the quantum transfer learning method, in different quantum real processors of IBM as well as in different simulators, in order to aid Coronavirus 2019 (COVID-19) detection by using a small number of computed tomography (CT) images as a diagnostic tool. Gokhale et al. (2020) present an extension to the quantum transfer learning approach formerly applied to image classification in order to solve the image splicing detection problem. Zen et al. (2020) proposed a method that evaluates the potential of transfer learning in order to improve the scalability of neural network quantum states. In the current era of intermediate-scale quantum technology, Mari’s method (Mari et al. 2019) is the most appealing as it allows optimal pre-processing of images with any state-of-the-art classical network and to process the most relevant features into a quantum computer.
2.3 Breast cancer screening using deep learning
The most popular datasets are the Mammography Image Analysis Society (MIAS) database with 322 image samples, the Digital Database for Screening Mammography (DDSM) containing 2500, and the Breast US Image with 250. Datasets can be used to extract regions of interest (ROIs) to perform detection and segmentation and ResNet variations as well as Inception, AlexNet, GoogleNet, and CaffeNet models have been used in breast cancer image analysis (Debelee et al. 2020).
In the survey by Debelee et al. (2020), we are presented with the available imaging modalities: screen-film mammography (SFM), digital mammography (DM), ultrasound (US), magnetic resonance imaging (MRI), digital breast tomosynthesis (DBT) or a combination of modalities. The digital image category has been the most effective and commonly used breast imaging modality despite having some limitations which include low specificity, leading to unnecessary biopsies.
An interesting work by Shen (2017) consists of an end-to-end training algorithm for whole-image mammograms projected to reduce the reliance on lesion annotations. Initially, it requires annotations to train the patch classifier consisting of ROI images, then a whole image classifier can be trained using only image-level labels. INbreast and DDSM datasets were used, showing that a whole image model trained on DDSM can be easily transferred to INbreast without using its lesion annotations and using a smaller amount of training data.
Studies, where transfer learning is used in breast cancer image analysis, can also be found; Mehra et al (2018) compared the results of three networks (VGG16, VGG19, and ResNet50) using BreakHis dataset, a dataset of histological breast cancer images, and applied data augmentation, using AUC, accuracy, and accuracy-precision score (APS) as performance measures. The pretrained networks were used as feature extractors and the extracted features were used to train logistic regression classifiers. A similar work was presented by Kassani et al. (2019). Also with a histological dataset, de Matos et al. (2019) used Inception-v3 as a feature extractor and a SVM classifier trained on a tissue labeled colorectal cancer dataset aiming to remove irrelevant patches. By doing so before training a second SVM classifier, this study shows that the accuracy improves when classifying malignant and benign tumors. Huynh et al. (2016) used data obtained from the University of Chicago Medical Center, consisting of 219 lesions on full-field digital mammography images and 607 ROIs about each lesion. By comparing SVM classifiers based on the CNN-extracted image features and their computer-extracted tumor features in the task of distinguishing between benign and malignant breast lesions this study concluded TL is a great tool when there are no large datasets. It is worth mentioning that Li et al. (2020) proposed a very interesting work building a fuzzy rule-based computer-aided diagnosis for mass classification of mammographic images using the Breast Cancer Data Repository (BCDR) dataset (Lopez et al. 2012).
In brief, the specificity and sensitivity of screening mammography are reported to be 89–97% and 77–87%, respectively. These metrics describe the performance of the models with reported false positive rates between 1 and 29% and sensitivities between 29 and 97% (Ribli et al. 2018). Regarding BCDR — specifically BCDR-D01 and BCDR-D02, the digital image datasets — using a CNN structure and by undersampling the dataset in order to balance both classes, Hepsaġ et al. (2017) obtained 62% accuracy, 75% training accuracy, 46% precision, 53% recall, and 51% F1-score. However, different results were achieved when selecting:
-
Masses: 88% test accuracy, 98% training accuracy, 86% precision, 90% recall, and F1-score of 88%,
-
Calcifications: 84%, training accuracy of 98%, precision of 91%, recall of 76%, and F1-score of 83%.
Cardoso et al. (2017) use BCDR in a different context, using ROI to perform a cross-sensor evaluation of mass segmentation methods. Diz et al. (2016) applied KNN, LibSVM, decision trees, random forest and naive Bayes to BCDR and achieved 89.3 to 64.7% for classifying each class benign/ malignant; 75.8 to 78.3% for classifying dense/fatty tissue and 71.0 to 83.1% for identification of a finding. Fontes et al. (2019) achieved the maximum accuracy of 76.9%, AUC of 74.9%, sensitivity of 84.88%, and specificity of 64.91% by applying a customized variant of Google’s InceptionV3 with the use of a Softmax classification layer as the output to perform the classification task.
However, there seems to be no studies where classical-quantum hybrid models are used to identify breast cancer in mammograms.
2.4 Transfer learning
Conventionally, deep neural networks need large amounts of labeled datasets and very powerful computing resources to solve challenging computer vision problems, e.g., feature extraction and classification. TL can be described as the improvement of learning a new task through the transfer of knowledge from a related (learned) task (Olivas 2009). This technique only works if the model features learned from the first task are general. It can be done using labeled or unlabeled data (Raina et al. 2007). From a pretrained model we can perform two types of tasks: (a) fine-tuning and (b) feature extraction.
In fine-tuning, we essentially retrain the model, updating most or all of the parameters having the pretrained weights as a basis. In feature extraction, we use the encoded features of the pretrained model to train the final layer’s weights (classifier) from which we derive predictions and reshape it to have the same number of outputs as the number of classes in the new dataset and freeze all the other layers, meaning that the pretrained CNN model works as a fixed feature extractor.
Transfer learning from natural image datasets, such as ImageNet, whose library is comprised of roughly one thousand classes (e.g., dog, car, or human), to medical imaging, such as chest pathology x-rays with about 5–14 classes (e.g., edema, pleural effusion, or cardiomegaly), is often used to avoid training the whole model (Morid et al. 2020).
3 Quantum machine learning
In quantum computing (QC), instead of working with binary digits (bits) we work with quantum bits, where 0 and 1 can overlap in time. The qubit (unit of quantum information) has two possible states |0〉 and |1〉. In the context of QC, the Hilbert space represents an abstract vector space which allows, e.g., a quantum superposition meaning that a physical system can be in more than one state simultaneously. The last few decades have seen significant advances in the fields of deep learning and quantum computing. As of today, huge amounts of data are being generated, pushing the interest surrounding the research at the junction of the two fields, leading to the development of quantum deep learning and quantum-inspired deep learning techniques. The upcoming topic is based on Schuld’s description (Schuld 2018).
3.1 Angle embedding
Angle embedding is an interesting approach in the context of neural networks. Assume a neural network and the network input 𝜃 = w0 + w1 ∗ x1 + ... + wN ∗ xN is written into the angle of an ancilla or net input qubit, which will be entangled with an output qubit in some arbitrary state |ψ〉, Ry(2v)|0〉⊗|ψ〉out, where v corresponds to an angle and Ry corresponds to the rotation around the y-axis, shown in Eq. 1:
Knowing this, we have what is also known as a quron in the context of quantum neural networks:
The next step is to prepare the output qubit Ry(2φ(v))|ψ〉, we can do this using a non-linear activation φ dependent on v that will rotate it.
3.2 QNN and variational circuits
A QNN is a machine learning model or algorithm that combines elements from quantum computing and artificial neural networks. Over the past decades, the term has been used to describe different ideas, ranging from quantum computers simulating the exact computations of neural nets, to general trainable quantum circuits that carry only little resemblance with the multi-layer perceptron structure. We will be focusing mainly on variational circuits since it is the one we will be using in this work, basing its description on the work of Mari et al. (2019). Increasingly, the term “quantum neural network” has been used to refer to variational or parameterized quantum circuits. Despite being mathematically different from the inner workings of neural networks, the term highlights the “modular” feature of quantum gates in a circuit, along with the use of tricks from training neural networks used in the optimization of quantum algorithms (Farhi and Neven 2018).
Classical-quantum hybrid approaches consisting of relatively low-depth quantum circuits called “variational algorithms” are the near term solution (Bharti et al. 2021; Cerezo et al. 2021; Mangini et al. 2021; Schuld et al. 2018). We can define a quantum layer as a unitary operation that can be processed by a low-depth variational circuit, producing the output state |y〉, by acting on the input state |x〉 of nq quantum subsystems (e.g., qubits or continuous variable modes).
where w is an array of classical variational parameters. A quantum layer could be, for example, a sequence of single-qubit rotations followed by a fixed sequence of entangling gates. Notice that, unlike a classical layer, a quantum layer retains the Hilbert space dimension of the input states. This fact is due to the fundamental unitary nature of quantum mechanics and should be taken into account when designing quantum networks.
A variational quantum circuit is a concatenation of q quantum layers, equivalent to the product of many unitaries parametrized by different weights:
A real vector x needs to be embedded into a quantum state |x〉 in order to enter a quantum network, this can also be done, depending on x, by a variational embedding layer and applied to some reference state (e.g., ground state).
The following step is to apply single-qubit rotations to x. The embedding layer \(\mathcal {E}\), unlike \({\mathscr{L}}\), maps from a classical vector space to a quantum Hilbert space. Conversely, the extraction of a classical output vector y from the quantum circuit can be obtained by measuring the expectation values of nq local observables \(\hat {y}=[\hat {y}_{1},\hat {y}_{2},...,\hat {y}_{n^{q}}]\). This process can be defined as a measurement layer, that maps a quantum state to a classical vector:
The full quantum network, including the initial embedding layer and the final measurement can be written as:
The full network maps a classical vector space to a classical vector space depending on classical weights. Despite containing a quantum computation hidden in the quantum circuit, \(\mathcal {F}\) is simply a black-box analogous to a classical deep network. However, there are technical limitations and physical constraints that should be taken into account: in a quantum network we do not have complete freedom when choosing the number of features in each layer, these numbers are often linked to the size of the physical system. Typically, embedding layers encode each classical element of x into a single subsystem where:
This can be overcome by:
-
1.
Adding ancillary subsystems and discarding or measuring some in the middle of the circuit,
-
2.
Engineering more complex embedding and measuring layers,
-
3.
Adding pre-processing and post-processing classical layers.
4 Experimental setup
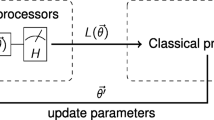
Our method consists of a classical-to-quantum TL scheme, based on the one proposed by Mari et al. (2019) where we assume two networks, A and B (Fig. 1):
General representation of the transfer learning method (Mari et al. 2019)
where \(A^{\prime }\) is the result of removing some of the final layers from network A, trained on dataset DA to perform task TA. \(A^{\prime }\) will be used as a feature extractor for B. B is the network that we want to train using the new dataset DB for some new task TB. In our problem, this can be translated to:
-
DA: ImageNet, image dataset with 1000 classes.
-
A: a pretrained ResNet18.
-
TA: classification, 1000 labels.
-
\(A^{\prime }\): a pretrained ResNet18 without the final linear layer, serving as an extractor of 512 features.
-
DB: BCDR, mammogram image dataset with 2 classes.
-
B: DressedQuantumNet, a dressed quantum circuit with 512 input features and 2 real outputs, proposed by Mari et al. (Mari et al. 2019).
-
TB: classification, 2 labels.
We decided it would be interesting to perform an initial comparison between different convolutional network architectures to verify which one best fits our data. Therefore, we chose some of the most widely used architectures in the literature. Using the previous scheme as reference, network A will be replaced by AlexNet (Krizhevsky et al. 2017), VGG19 (Simonyan and Zisserman 2014), DenseNet161 (Huang et al. 2017), and ResNeXt50 (Xie et al. 2017) for the experiments. Further experiments will be done using the one with the best performance.
4.1 Dressed quantum circuit
The dressed quantum circuit (DressedQuantumNet) consists of the classifier that will attach to the final linear layer of the pretrained model.
Assuming a Resnet18 is the pretrained model, the classifier will receive 512 real values as input to the circuit and will output 2 real values. The only trainable part of the network is the quantum classifier. Therefore, the number of trainable parameters can be calculated using the circuit’s input size, its depth, and output size: 512 ∗ n_qubits + n_qubits ∗ depth + output_size. Trainable parameters’ count will be presented along our results. The following represents the dressed quantum circuit:
where \({\mathscr{L}}_{512 \xrightarrow {} 4}\) is a pre-processing layer that consists of an affine operation followed by a non-linear function φ = tanh applied element-wise, Q is the variational circuit and \({\mathscr{L}}_{4 \xrightarrow {} 2}\) is a linear classical layer without activation (i.e., φ(y) = y).
The 4 real variables obtained from \({\mathscr{L}}_{512 \xrightarrow {} 4}\) are then embedded in the quantum circuit by applying a Hadamard gate (H) and performing a rotation around the y-axis of the Bloch sphere parametrized by a classical vector x:
The trainable circuit is composed of q variational layers \(\mathcal {Q} = {\mathscr{L}}_{q} \circ ... \circ {\mathscr{L}}_{2} \circ {\mathscr{L}}_{1}\) where:
where K is an entangling unitary operation made of three controlled NOT gates.
Finally, the 4 output states are measured on the classical register using the Pauli-Z matrix and passed to the linear classical layer, producing 2 output states. The classification is done according to argmax(y), where y = (y1,y2) is the output of the dressed quantum circuit. An illustration of the circuit is shown in Fig. 2.
4.2 Overview
The implementation for this research was based on the one provided by Mari et al. (2019). Concerning the data, it was loaded using our own dataloader (BCDR), and transformed using random horizontal and vertical flips for data augmentation. The input channels are also normalized using mean values and standard deviations of ImageNet. In order to get reproducible results, a manual seed was set for the dataloader.
Hybrid classical-quantum model
We start by loading a pretrained model and managed the following considerations:
-
1.
Since we want to use the pretrained network as a feature extractor, we need to freeze every layer so that the weights are not updated during training, the only weights we will be updating are the classifiers.
-
2.
We determine the size of the vector that will be entering DressedQuantumNet and set DressedQuantumNet as the model’s classifier layer.
-
3.
Cross-entropy was used as a loss function, Adam optimizer was selected to update the weights of the model at each training step, and a scheduler was set to decay the learning rate by gamma every step size.
-
4.
Finally, we train the model.
5 Materials, tests and results
Model comparison tests were performed, classical without and with TL, also comparison using a quantum simulator and resorting to a quantum device available at IBM Quantum (https://quantum-computing.ibm.com/).
In this section we give a description of the data used and how it was handled, followed by testing, analysis and discussion of the obtained results. Tests were run on Intel Core i7-10510U CPU @ 1.80GHz×8 with 16GB RAM. The quantum experiments were conducted using the IBM Quantum device ibm_lagos which is detailed later.
5.1 Materials, data description and processing
In this section we briefly describe the data and the needed processing for the data preparation in order to attain the achieved results. We used RStudio in this portion of our work. The data used was provided by BCDR (Lopez et al. 2012). According to their website (Guevara 2020), “the creation of BCDR was supported by the IMED Project (for Development of Algorithms for Medical Image Analysis) aimed at creating medical image repositories and massive exploration of Computer-Aided Diagnosis (CADx) methods on GRID computing resources. The IMED project was carried out by INEGI, FMUP-CHSJ – University of Porto, Portugal and CETA-CIEMAT, Spain between March 2009 and March 2013. Recently, in October of 2013, the IMED project was renewed and Aveiro University began to be part of this consortium. Now, the four institutions continue to actively augment and develop the BCDR.”
The data is composed of two folders (BCDR_D01 and BCDR_D02) divided by study and patient, each containing several files corresponding to cranio-caudal (CC) and medium-lateral oblique (O) views of one or both sides — left (L) and right (R) — of a breast. The data also includes information, such as, the biopsy result. This new dataframe is composed of 49 columns and 2696 rows. From image filename we cropped out image view and side and created a new column view. The new column view was filled according to the following: LCC = 1, LO = 2, RCC = 3 and RO = 4.
When looking for columns with at least 30% of NA values, we also made sure to trim every value since there were some white spaces in the dataset. Variables with a single class were also eliminated. Columns with 2 to 10 different values were converted to factor. We did the same for rows, eliminating those with at least 30% of NA values, no rows were found. Column classification had a row with value 2, this row was eliminated. Finally, we changed classification class Malignant to 1 and Benign to 0 and applied the unique function. The resulting dataframe contains 909 rows and 10 columns. Note that the same mammogram can contain the description of several different masses/calcifications. Knowing this, 269 mammograms contain nodules, 688 contain calcifications, 82 contain microcalcifications, 8 contain architectural distortions and 51 contain stroma distortions. Since we will be analyzing unique images, we eliminate duplicated image filename from the dataset, and end up with 825 rows.
To check if the images are well represented, we analyzed features age, view, density and classification. Figure 3 shows data distribution of the features age, view, density and classification.
As we can see, classification is unbalanced. Class 1 has 141 samples and class 0 has 684. We fixed this by oversampling class 1 by adding 2 copies of every sample with classification = 1 and cropping the excess samples with classification = 0. Each set of copies will be associated with the angle by which it will be rotated. We chose angle 60 for the first set of copies, and angle 45 for the second set. Since every image is subjected to random horizontal and vertical flips during train and test phases, we kept the angles to the first quadrant such that they do not result in the same images. The distribution, after balancing classification, can be seen below.
Figure 4 shows data distribution of the features age, view, density and classification after balancing the feature classification.
5.2 Tests and results
In this chapter, we present:
-
An initial model comparison using a classical approach in order to determine which of the models has the best performance. We also determine the proper learning rate for each of the models that will be further tested;
-
Tests and results using our QuantumDressedNetwork as a classifier resourcing to a quantum simulator in order to pick which hybrid classical-quantum model will be further tested;
-
Tests and results using a classical model without and with TL to benchmark against our hybrid model. Then a comparison between results obtained from these models;
-
Tests and results using our QuantumDressedNetwork as a classifier resourcing to a real quantum device.
Python and PennyLane platform were used in this portion of our work. Heatmaps which explicitly model the contributions of each pixel in the feature maps of a CNN to the final output were used to aid result comparison. These heatmaps result from using GradCAM (Selvaraju et al. 2017), an earlier gradient-based visual explanation method, and GradCAM++ (Chattopadhay et al. 2018). Note that these networks have different architectures and, therefore, the final convolutional layer, the one evaluated by GradCAM and GradCAM++, is different. Some of the functions were deprecated and were therefore changed manually.
We defined class 0 as the negative class as it represents a benign mammogram, and class 1 as a positive class as it represents a malignant mammogram.
In Table 1 we can observe the values of the parameters for the different approaches, where, step is the step of the learning rate, # epochs is the number of training epochs, # batch size is the number of samples for each training step, gamma lr scheduler is the learning rate reduction applied every 10 epochs and step size concerns the learning rate updates every step size epochs. Model comparison corresponds to the initial comparison of all the models and Classical to the parameters set in the classical without and with TL.
Model comparison
A positive case and a negative case were tested against AlexNet, VGG19, DenseNet161, ResNet18, and ResNeXt50_32x4d respectively. For simplicity, we used a simple linear transformation as classifier.
In Fig. 5, the warmer the color, the more the network is targeting the area. Knowing this, DenseNet, Resnet, and ResNeXt seem to do a pretty good job at targeting the main areas where masses/calcifications are present.
The rows in (a) and (b) show the results from AlexNet, VGG19, DenseNet161, ResNet18, and ResNeXt50_32x4d, respectively. The first column corresponds to the original image, the following two result from GradCAM (without image overlap and with image overlap) and the last two result from GradCAM++ (without image overlap and with image overlap)
Now we need to have an idea of which learning rate to use. The starting learning rate is 0.4 and will be adjusted by the model by 10-1 every 5 epochs. The goal is to find the optimal learning rate through the graphic, by choosing the global minimum, as suggested in the paper by Smith (2017) so we can give every model a fair chance to converge.
Table 2 shows the training results for DenseNet, ResNet and ResNeXt.
The best learning rate should be the value that is approximately in the middle of the sharpest downward slope. Therefore, we have that DenseNet’s, ResNet’s, and ResNeXt’s optimal learning rate is around 0.0004. We will keep step_size = 10 because TL requires small learning rates regardless.
Quantum simulator
Using the best parameters mentioned we trained and tested DenseNet, ResNet and ResNext in a quantum simulator, with the QuantumDressedNetwork as a classifier for Resnet. We also tune the last parameter needed; the circuit’s depth. According to Mari et al. (2019), a characteristic of the TL approach is the existence of an intermediate optimal value for the quantum depth.
Table 3 shows the performance metrics results for DenseNet, ResNet and ResNeXt, in the quantum simulator. Results are very similar for all three models. However, ResNet achieved a considerably higher recall and AUC. Based on this, we will choose ResNet. Additionally, it is faster in comparison. Figure 6 shows results varying the circuit depth from 1 to 4.
The rows in (a), (b), (c), and (d) show the results for a positive and for a negative sample, respectively, using Resnet. The first column corresponds to the original image, the following two result from GradCAM (without image overlap and with image overlap) and the last two result from GradCAM++ (without image overlap and with image overlap)
Classical (without and with) TL
Figure 7a shows classical results without TL obtained from a ResNet, meaning that the network was trained from scratch. Figure 7b shows classical results with TL obtained from a ResNet, meaning that the only trained layers were the classifier’s. These will be used as benchmarks to our model. For simplicity, we used a simple linear transformation as classifier.
The rows in (a) and (b) show the results for a positive and for a negative sample, respectively. The first column corresponds to the original image, the following two result from GradCAM (without image overlap and with image overlap) and the last two result from GradCAM++ (without image overlap and with image overlap)
Let’s consider the models:
-
C: the classical ResNet.
-
C+TL: the classical ResNet using transfer learning.
-
CQ1, CQ2, CQ3 and CQ4: the hybrid classical-quantum ResNet models using transfer learning, whose variational quantum circuits have a depth of 1, 2, 3, and 4, respectively (using the simulator).
As mentioned before, performance metrics are calculated considering that the class malignant is the positive class (Table 4).
CQ1 has the higher overall results between training and test, namely precision, specificity and AUC score. It also has the lowest loss value. No further testing was needed on depth as the AUC value started to degrade. The resulting model is used on an IBM Quantum device for testing.
Quantum device
Due to the waiting times, IBM Quantum experiments were done using the best model state that resulted from training in the simulator. This means that only the test part was done in the quantum machine. We ran the file containing our network (that resulted from the dressed quantum circuit with depth = 1 — CQ1 — in the state where the best F1-score was measured), on an IBM Quantum device.
Figure 8 shows the quantum results achieved for the metrics under consideration resorting to the device ibm_lagos, for 20 repetitions of the testing in the same testset. As, in this experiment, we use data augmentation for each run, results are not the same at each repetition, but the model seems to be stable.
In Table 5 we show the metrics results for the best classification given by the quantum device among the 20 repetitions.
At first, we believed that the testing results variation seen in Fig. 8a originated from the quantum machine error rates. The configuration of the qubits connectivity showing associated errors of the quantum device we use, ibm_lagos, is shown in Fig. 9. The main characteristics of this device are as follows: 7 qubits, 32 of quantum volume, a Falcon r5.11H processor type, version 1.0.1, the basis gates (CX, ID, RZ, SX, X), 6.595e-3 of average CNOT error, 1.466e-2 of average readout error, 106.39 us average T1 and 79.45 us average T2. The readout error of each qubit is shown on the left hand side. Errors can be as high as 2.53%. The average error for Hadamard and CNOT gates are also shown. As the errors are not too high to justify changes in performance results, we performed an experiment without data augmentation.
Using the resulting model from CQ1 as a base, and without performing any data augmentation techniques, we tested the same exact 32 samples (the size of one epoch):
-
once in the quantum simulator;
-
twice on ibmq_quito (due to queue size).
The results were exactly the same:
-
6 of our samples were positive but incorrectly classified as negative;
-
26 of our samples were negative and correctly classified as negative.
As expected, the results were the same for testing of the same samples. This leads us to believe that noise has little or no interference in our model and that the quantum simulator does a good job of simulating a quantum device.
6 Conclusion and future work
It is too soon to say whether or not QML can be an advantage. Our results only point to a good generalization of complex data which needs further testing. The classical residual neural network, without resourcing to TL, achieved a maximum of 67% accuracy while our other experiments using TL achieved 84%. When comparing classical ResNet with and without applying TL method we can see that the task execution time was cut by half. The results are not so drastic when comparing the classical-classical and the classical-quantum hybrid networks making use of TL. The results for every metric are far more consistent between training and testing and between epochs when using the dressed quantum circuit, so much so that AUC value went from 58 to 77%, leading us to believe that a hybrid-quantum network does present some advantage as AUC measures how well the model separates two classes. Our overall results surpassed those from the literature that also applied a classical transfer learning technique — accuracy of 84% against 76.9%, AUC of 77% against 74.9% and specificity of 100% surpassed 64.91%. However, regarding recall we obtained 69% and 84.88% was mentioned in the literature. Nevertheless, the results from the quantum device show a recall closer to that of the literature, 81%.
As is known in the community, there has been a lot of work in the field of error correction regarding quantum machines. However, despite the errors associated to the used quantum device ibm_lagos, good results were obtained with the quantum experiments. Our results seem to confirm what has been said in the quantum machine learning literature: the results seem to converge towards an optimal solution much earlier, which leads us to believe that such a large number of iterations in the quantum version will not be needed compared to the classical version.
As future work, an interesting approach could be to separate the dataset into masses and calcifications and only then perform classification. That way, we would be evaluating different specific features in a similar scenario and would probably get clearer results. It would also be interesting to test different datasets. We also would like to explore other gate-based quantum devices with smaller associated errors. Lastly, the best way to know how well this model performs would be to see how the data appears in our Hilbert space. Ideally, both classes would appear very separately in it, as tight clusters.
In addition, in this research we conduct the experiments using the angle embedding technique since with this method the features can be learned. The same is not true if we use, for example, the basis embedding because it is theoretically impossible. In addition, if we had used, e.g., the amplitude embedding, it could have been too complex to compute gradients with respect to features, which may not be feasible. However, in the future other embeddings can be considered (Lloyd et al. 2020).
References
Acar E, Yilmaz I (2020) Covid-19 detection on ibm quantum computer with classical-quantum transfer learning. medRxiv
Bharti K, Cervera-Lierta A, Kyaw TH, Haug T, Alperin-Lea S, Anand A, Degroote M, Heimonen H, Kottmann JS, Menke T, Mok WK, Sim S, Kwek LC, Aspuru-Guzik A (2021) Noisy intermediate-scale quantum (nisq) algorithms. arXiv:2101.08448
Biamonte J, Wittek P, Pancotti N, Rebentrost P, Wiebe N, Lloyd S (2017) Quantum machine learning. Nature 549(7671):195
Cardoso JS, Marques N, Dhungel N, Carneiro G, Bradley AP (2017) Mass segmentation in mammograms: a cross-sensor comparison of deep and tailored features. In: 2017 IEEE International Conference on Image Processing (ICIP). IEEE, pp 1737–1741
Cerezo M, Arrasmith A, Rea B (2021) Variational quantum algorithms. Nat Rev Phys 3:625–644. https://doi.org/10.1038/s42254-021-00348-9
Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN (2018) Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE, pp 839–847
Chen S, Ma K, Zheng Y (2019) Med3d: Transfer learning for 3d medical image analysis. arXiv:190400625
Debelee TG, Schwenker F, Ibenthal A, Yohannes D (2020) Survey of deep learning in breast cancer image analysis. Evolving Systems 11(1):143–163
Deutsch D (1985) Quantum theory, the church–turing principle and the universal quantum computer. Proceedings of the Royal Society of London A Mathematical and Physical Sciences 400(1818):97–117
Diz J, Marreiros G, Freitas A (2016) Applying data mining techniques to improve breast cancer diagnosis. Journal of Medical Systems 40(9):203
Duan B, Yuan J, Xu J, Li D (2019) Quantum algorithm and quantum circuit for a-optimal projection: Dimensionality reduction. Physical Review A 99(3):032311
Farhi E, Neven H (2018) Classification with quantum neural networks on near term processors. arXiv:180206002
Feynman RP (1982) Simulating physics with computers. International Journal of Theoretical Physics 21(6):467–488
Fontes JP, Lopez MG et al (2019) Representation learning approach to breast cancer diagnosis. In: European Congress of Radiology-ECR, p 2019
Fund WCR (2020) Breast cancer: How diet, nutrition and physical activity affect breast cancer risk. https://www.wcrf.org/dietandcancer/breast-cancer
Gao W, Zhang X, Yang L, Liu H (2010) An improved sobel edge detection. In: 2010 3Rd international conference on computer science and information technology, vol 5. IEEE, pp 67–71
Gokhale A, Pande MB, Pramod D (2020) Implementation of a quantum transfer learning approach to image splicing detection. International Journal of Quantum Information 18(05):2050024
Grant E, Benedetti M, Cao S, Hallam A, Lockhart J, Stojevic V, Green AG, Severini S (2018) Hierarchical quantum classifiers. npj Quantum Information 4(1):1–8
Grover LK (1996) A fast quantum mechanical algorithm for database search. In: Proceedings of the twenty-eighth annual ACM symposium on Theory of computing, pp 212–219
Guevara MA (2020) Breast cancer digital repository. https://bcdr.eu/information/about
Havlíček V, Córcoles AD, Temme K, Harrow AW, Kandala A, Chow JM, Gambetta JM (2019) Supervised learning with quantum-enhanced feature spaces. Nature 567(7747):209
Henderson M, Shakya S, Pradhan S, Cook T (2020) Quanvolutional neural networks: powering image recognition with quantum circuits. Quantum Machine Intelligence 2(1):1–9
Hepsaġ PU, Özel SA, Yazıcı A (2017) Using deep learning for mammography classification. In: 2017 International conference on computer science and engineering (UBMK). IEEE, pp 418–423
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Huynh BQ, Li H, Giger ML (2016) Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of Medical Imaging 3(3):034501
Kassani SH, Kassani PH, Wesolowski MJ, Schneider KA, Deters R (2019) Breast cancer diagnosis with transfer learning and global pooling. arXiv:190911839
Kaur R, Sharma R, Kumar P (2018) An efficient speaker recognition using quantum neural network. Modern Physics Letters B 32(31):1850384
Kerenidis I, Landman J, Prakash A (2019) Quantum algorithms for deep convolutional neural networks. arXiv:191101117
Kim HG, Choi Y, Ro YM (2017) Modality-bridge transfer learning for medical image classification. In: 2017 10Th international congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI). IEEE, pp 1–5
Krishna CR, Dutta M, Kumar R (2018) Proceedings of 2nd International Conference on Communication, Computing and Networking: ICCCN 2018, NITTTR Chandigarh, vol 46. Springer, India
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Lévy D, Jain A (2016) Breast mass classification from mammograms using deep convolutional neural networks. arXiv:161200542
Li F, Shang C, Li Y, Shen Q (2020) Interpretable mammographic mass classification with fuzzy interpolative reasoning. Knowl-Based Syst 191:105279
Liu Y, Zhang X, Cai G, Chen Y, Yun Z, Feng Q, Yang W (2019) Automatic delineation of ribs and clavicles in chest radiographs using fully convolutional densenets. Computer Methods and Programs in Biomedicine 180:105014
Lloyd S, Schuld M, Ijaz A, Izaac J, Killoran N (2020) Quantum embeddings for machine learning. arXiv:2001.03622
Lopez MG, Posada N, Moura DC, Pollán R R, Valiente JMF, Ortega CS, Solar M, Diaz-Herrero G, Ramos I, Loureiro J et al (2012) BCDR: a breast cancer digital repository. In: 15Th international conference on experimental mechanics, vol 1215
Lu S, Lu Z, Zhang YD (2019) Pathological brain detection based on alexnet and transfer learning. Journal of Computational Science 30:41–47
Mangini S, Tacchino F, Gerace D, Bajoni D, Macchiavello C (2021) Quantum computing models for artificial neural networks. IOPScience 134(1):10002. https://doi.org/10.1209/0295-5075/134/10002
Mari A, Bromley TR, Izaac J, Schuld M, Killoran N (2019) Transfer learning in hybrid classical-quantum neural networks. arXiv:191208278
de Matos J, Britto ADS, Oliveira LE, Koerich AL (2019) Double transfer learning for breast cancer histopathologic image classification. In: 2019 International joint conference on neural networks (IJCNN). IEEE, pp 1–8
Mehra R et al (2018) Breast cancer histology images classification: Training from scratch or transfer learning? ICT Express 4(4):247–254
Morid MA, Borjali A, Del Fiol G (2020) A scoping review of transfer learning research on medical image analysis using imagenet. arXiv:200413175
Nunnari F, Bhuvaneshwara C, Ezema AO, Sonntag D (2020) A study on the fusion of pixels and patient metadata in cnn-based classification of skin lesion images. In: International cross-domain conference for machine learning and knowledge extraction. Springer, pp 191–208
Olivas ES (2009) Handbook of research on machine learning applications and trends: Algorithms, methods and techniques: Algorithms, Methods, and Techniques. IGI Global
Omonigho EL, David M, Adejo A, Aliyu S (2020) Breast Cancer: Tumor detection in mammogram images using modified alexnet deep convolution neural network. In: 2020 International conference in mathematics, computer engineering and computer science (ICMCECS). IEEE, pp 1–6
Preskill J (2018) Quantum computing in the nisq era and beyond. Quantum 2:79
Raina R, Battle A, Lee H, Packer B, Ng AY (2007) Self-taught learning: transfer learning from unlabeled data. In: Proceedings of the 24th international conference on Machine learning, pp 759–766
Ribli D, Horváth A, Unger Z, Pollner P, Csabai I (2018) Detecting and classifying lesions in mammograms with deep learning. Scientific Reports 8(1):1–7
Sang J, Wang S, Li Q (2017) A novel quantum representation of color digital images. Quantum Inf Process 16(2):42
Schuld M (2018) Supervised learning with quantum computers. Springer, Berlin
Schuld M, Bocharov A, Svore K, Wiebe N (2018) Circuit-centric quantum classifiers. arXiv:180400633
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
Shen L (2017) End-to-end training for whole image breast cancer diagnosis using an all convolutional design. arXiv:171105775
Shie CK, Chuang CH, Chou CN, Wu MH, Chang EY (2015) Transfer representation learning for medical image analysis. In: 2015 37Th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 711–714
Shor PW (1999) Polynomial-time algorithms for prime factorization and discrete logarithms on a quantum computer. SIAM Review 41(2):303–332
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:14091556
Skolik A, McClean JR, Mohseni M, van der Smagt P, Leib M (2020) Layerwise learning for quantum neural networks. arXiv:200614904
Smith LN (2017) Cyclical learning rates for training neural networks. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE, pp 464–472
Tang X, Shu L (2014) Classification of electrocardiogram signals with rs and quantum neural networks. International Journal of Multimedia and Ubiquitous Engineering 9(2):363–372
Wang SH, Xie S, Chen X, Guttery DS, Tang C, Sun J, Zhang YD (2019) Alcoholism identification based on an alexnet transfer learning model. Frontiers in Psychiatry 10:205
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1492–1500
Yan F, Huang X, Yao Y, Lu M, Li M (2019) Combining lstm and densenet for automatic annotation and classification of chest x-ray images. IEEE Access 7:74181–74189
Zen R, My L, Tan R, Hébert F, Gattobigio M, Miniatura C, Poletti D, Bressan S (2020) Transfer learning for scalability of neural-network quantum states. Physical Review E 101(5):053301
Zhang K, Hsieh MH, Liu L, Tao D (2020) Toward trainability of quantum neural networks. arXiv:201106258
Zhang L, Yang G, Ye X (2019) Automatic skin lesion segmentation by coupling deep fully convolutional networks and shallow network with textons. Journal of Medical Imaging 6(2):024001
Zhang Y, Lu K, Xu K, Gao Y, Wilson R (2015) Local feature point extraction for quantum images. Quantum Inf Process 14(5):1573–1588
Acknowledgements
We would like to thank Prof. Peter Wittek’s (in memoriam) for his valuable comments concerning variational circuits and how a bridge between gate-based quantum computing and the adiabatic paradigm are made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors Vanda Azevedo and Carla Silva contributed to this research equally.
Rights and permissions
About this article
Cite this article
Azevedo, V., Silva, C. & Dutra, I. Quantum transfer learning for breast cancer detection. Quantum Mach. Intell. 4, 5 (2022). https://doi.org/10.1007/s42484-022-00062-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42484-022-00062-4