Abstract
Due to its high infectivity, COVID-19 has rapidly spread worldwide, emerging as one of the most severe and urgent diseases faced by the global community in recent years. Currently, deep learning-based diagnostic methods can automatically detect COVID-19 cases from chest X-ray images. However, these methods often rely on large-scale labeled datasets. To address this limitation, we propose a novel neural network model called CN2A-CapsNet, aiming to enhance the automatic diagnosis of COVID-19 in chest X-ray images through efficient feature extraction techniques. Specifically, we combine CNN with an attention mechanism to form the CN2A model, which efficiently mines relevant information from chest X-ray images. Additionally, we incorporate capsule networks to leverage their ability to understand spatial information, ultimately achieving efficient feature extraction. Through validation on a publicly available chest X-ray image dataset, our model achieved a 98.54% accuracy and a 99.01% recall rate in the binary classification task (COVID-19/Normal) on a six-fold cross-validation dataset. In the three-class classification task (COVID-19/Pneumonia/Normal), it attained a 96.71% accuracy and a 98.34% recall rate. Compared to the previous state-of-the-art models, CN2A-CapsNet exhibits notable advantages in diagnosing COVID-19 cases, specifically achieving a high recall rate even with small-scale datasets.
Article Highlights
-
Developing a novel deep learning model for assisting doctors in automating the screening of COVID-19 cases.
-
This study has opened up new directions for improving the accuracy of COVID-19 diagnosis.
-
Achieving high recall rate in diagnosis with limited chest X-ray images.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In 2020, the novel coronavirus (COVID-19) was swiftly disseminated worldwide. This coronavirus typically triggers symptoms of fever and pneumonia and exhibits distinct person-to-person transmission characteristics and high infectivity [1, 2]. Vulnerable populations, particularly the elderly, children, individuals with weakened immune systems, and those with underlying health conditions, face a heightened risk of infection [3]. As of August 27, 2023, the global tally has reported over 770 million confirmed cases and more than 6.9 million fatalities [4]. Owing to the virus’s exceptional transmissibility, many nations’ healthcare systems confront unprecedented strains, with some even experiencing states of paralysis. Consequently, timely detection and isolation constitute pivotal measures for halting the pandemic’s progression, and policies advocating social distancing prove effective in curtailing the rapid dissemination of the virus [5]. The coronavirus’s impact on lung tissue can be visually discerned through medical imaging, with apparent correlations between radiological manifestations [6]. Therefore, widespread screening becomes an imperative mandate to detect and impede viral transmission within patients. To mitigate the infection risk associated with nucleic acid collection, computer-aided diagnosis has seen extensive application in virus detection, facilitating the automation of the testing process. This, in turn, alleviates the burden on healthcare professionals and enhances the efficiency of epidemic prevention efforts.
X-ray images, in comparison to CT scans, are more cost-effective, quicker to obtain, and involve lower radiation exposure. However, manual pneumonia diagnosis is a challenging task. Experts must meticulously inspect and interpret the white spots in X-ray images [7]. Furthermore, pneumonia with a speckled pattern can be easily misdiagnosed as tuberculosis or bronchiectasis [8]. To address this issue, Fei Shan introduced a deep learning-based segmentation system designed to automatically quantify the proportion between regions of interest and lung volume. This system’s performance was validated through a comparison involving the automatic segmentation of infected areas and the manual annotation of infected regions in 300 chest CT scans from 300 COVID-19 patients [9]. Furthermore, Nilanjan Dey utilized traditional convolutional neural networks, including models such as VGG19, VGG16, AlexNet, and ResNet50, to identify patients’ X-ray images. Among these models, VGG19 was demonstrated to be the most effective. This experiment further reaffirmed the feasibility of employing convolutional neural networks for COVID-19 diagnosis using X-ray images [10,11,12,13]. With the continuous improvement of deep learning models and the increasing richness of datasets, the evaluation of large-scale datasets has become more convenient. For example, Alshmrani et al. [14] collected a total of 80,000 images and employed their proposed VGG19+CNN model for a six-class classification task. The research results, evaluated based on metrics such as accuracy, recall, and F1 score, demonstrated an accuracy of 96.48%, a recall rate of 93.75%, and an F1 score of 95.62%. Moreover, to address the issues of limited publicly available COVID-19 datasets and the significant class imbalance in multi-class classification tasks, Roy et al. [15] introduced an augmentation technique named SVD-CLAHE. They leveraged ResNet-50 on this augmented dataset and achieved superior results compared to other models. Bilal et al. [16] employed ABC optimization techniques for image enhancement, leading to a diagnostic accuracy of 98.97% for their combined ResNet-101 and LSTM model, an improvement of 1.16% over the original model. Additionally, Lee et al. [17] performed data augmentation on top of DenseNet201 and fine-tuned the final layers to overcome the issue of limited COVID-19 samples and the resulting data scarcity.
In the field of medicine, convolutional neural networks (CNNs) are currently the most popular approach for deep learning [18,19,20]. However, CNN models come with certain constraints, one of which is the max-pooling operation. Max-pooling is designed to pass the most valuable information from the previous layer to the next, but this can result in the loss of fine details in the data, and the data might not be transmitted to other layers effectively. Moreover, existing CNN models fail to maintain the part-whole relationships of objects. To overcome the limitations of CNNs, Sabour et al. introduced a novel neural network architecture in 2017, known as capsule networks [21]. They claimed that the proposed model overcomes the drawbacks of existing CNN models. In a study on capsule networks and X-ray images, Lobna et al. [22] optimized the capsule network parameters using Gaussian optimization methods, achieving an accuracy of 96.58% on an imbalanced dataset. Unlike traditional CNNs that use scalar models, capsule networks represent the position of each unit in the image using vectors, thus preserving the relative spatial relationship information between elements that might be lost due to max-pooling. In our research, leveraging the characteristics of capsule networks proved to be an effective strategy.
Chest X-ray images play a crucial role in medical diagnosis. However, effectively utilizing these image features is a key challenge in diagnosis due to redundancies and correlations. Currently, many methods rely on large-scale datasets or data augmentation techniques to enhance diagnostic accuracy. Training with large datasets can yield high accuracy, but it requires extended training times and demands a substantial quantity of data. On the other hand, data augmentation techniques can improve model performance on limited datasets, but excessive reliance on similar samples may reduce the model’s generalization ability. Therefore, finding a balance between utilizing the limited features in chest X-ray images to ensure accuracy and enhancing the model’s generalizability is a challenge in the field of chest X-ray image diagnosis. To address this issue, this study proposes a method that combines capsule networks and convolutional neural networks (CNNs), incorporating the Convolutional Block Attention Module (CBAM) to enhance the model’s focus on specific regions of X-ray images and improve the representation capability of these regions. The capsule network captures correlations between different regions in chest X-ray images, enabling better utilization of image features. The CBAM module further strengthens the CNN’s focus on essential areas, thereby enhancing the model’s performance. Utilizing the method proposed in this study achieves high recall rates on limited datasets and demonstrates excellent diagnostic results. Compared to traditional methods that rely on large datasets or data augmentation techniques, this approach maintains accuracy while offering higher computational efficiency and better generalization capabilities. Hence, this research provides valuable guidance on how to effectively utilize chest X-ray image features and improve diagnostic accuracy in situations with limited datasets.
In our study, to deliver a more objective assessment of our model, we validated it using a publicly available dataset. The main contributions are as follows:
-
(1)
CN2A-CapsNet combines the spatial understanding capability of capsule networks to address the issue of relative spatial relationship information loss in traditional deep learning models. Through experiments, we discovered that the relative spatial positional information in chest X-ray images is an important basis for diagnosing COVID-19 patients.
-
(2)
By integrating CNN with attention mechanisms, the proposed CN2A model can adaptively determine weights, enabling the network to efficiently explore relevant information and feed relevant features into the capsule network, thus fully leveraging the capsule network’s sensitivity to relative spatial relationships.
-
(3)
Compared to methods proposed by other researchers, CN2A-CapsNet achieves high recall rates even with small-scale datasets, making it more suitable for practical application scenarios.
The subsequent sections of this article are organized as follows: Sect. 2 provides a detailed description of the proposed COVID-19 diagnostic method, Sect. 3 discusses the specific procedures and results of two main experiments, as well as comparisons with methods proposed by other researchers. Section 4 provides a discussion of the study, and Sect. 5 summarizes the conclusions of the research.
2 Materials and methods
2.1 Data collection
This study is based on a publicly available large Chest X-ray (CXR) image dataset [23, 24], which was collaboratively created by researchers from Hamad Medical Corporation in Qatar, Dhaka University in Bangladesh, and collaborators and medical practitioners from Pakistan and Malaysia. The dataset comprises 33,920 chest X-ray images categorized into three classes: 11,956 COVID-19 positive cases, 11,263 non-COVID-19 infections (viral or bacterial pneumonia), and 10,701 images depicting normal conditions. We conducted experiments with two categories (COVID-19/Normal) and three categories (COVID-19/Pneumonia/Normal) using this dataset, as illustrated in Fig. 1. To mitigate the impact of data bias and ensure a more objective model evaluation, we randomly selected 2,400 images from the dataset and split them into training and testing sets in a 5:1 ratio. Furthermore, the original images in the dataset are of high resolution. To standardize the processing, we resized them to grayscale images of 299\(\times \)299 pixels and normalized the pixel values from the [0, 255] range to [0, 1].
2.2 Networks
2.2.1 VGG network
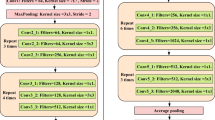
In 2014, Oxford University and Google collaborated to develop VGG, a deep convolutional neural network. This network investigated the relationship between performance and depth and successfully constructed network architectures with 16 to 19 layers. The research findings demonstrated that increasing the depth of the network can improve its performance to a certain extent, significantly reducing the error rate. Furthermore, the network exhibited good scalability and generalization capabilities, enabling it to adapt to different image datasets [25].
VGG19 is a convolutional neural network model that consists of 16 convolutional layers and 3 fully connected layers. In the traditional VGG19 architecture, the model is used for performing a 10-class classification task. However, in this study, we modified the fully connected layers to output 2 class labels. The structure of VGG19 is illustrated in Fig. 2.
2.2.2 Capsule network
The capsule network, proposed by Hinton in 2017 [21], consists of two main components: PrimaryCaps and DigitCaps. Each capsule unit in the network contains multiple neurons. Unlike traditional CNNs that use scalar outputs, capsule networks use vectors to represent the position of each unit in the image, thereby preserving the relative spatial relationship information between elements that may be lost in traditional CNNs due to max pooling. Additionally, capsule networks introduce a dynamic routing process to update parameters. Unlike max pooling, this process does not discard precise positional information.
The capsule network uses Squashing as its activation function, as shown in Eq. (1):
In the equation, \(\nu _j\) represents the output of the capsule, and \(s_j\) represents the total input to the capsule.
Another crucial component of capsule networks is the dynamic routing process. Within a capsule, the coupling coefficients \(c_{ij}\) are iteratively updated using the dynamic routing algorithm to compute the output of the capsule, as described in Eq. (2).
where \(b_{ij}\) represents the dynamic weight from the i th capsule to the j th capsule, which is updated through the iterative dynamic routing algorithm to obtain \(c_{ij}\). The updated coupling coefficient \(c_{ij}\) and the predicted vector \(\hat{u}_{j|i}\) are then weighted summed to obtain \(s_j \), as shown in Eq. (3). The predicted vector \(\hat{u}_{j|i}\) is obtained by multiplying the transformation matrix \(W_{ij}\) with the output \(u_i\) of the preceding layer, as depicted in Eq. (4).
In the capsule network proposed by Hinton, the classical capsule network consists of a convolutional layer followed by two capsule layers. The network employs a margin loss function \(L_c\) as the sole loss function, as shown in Eq. (5).
The Capsule Network consists of two capsule layers: the PrimaryCaps and the DigitCaps, as shown in Fig. 3. In the PrimaryCaps layer, the feature maps are reconstructed and 32 capsules are generated. In the DigitCaps layer, there are 10-digit capsules, with each digit capsule corresponding to a specific digit category. The DigitCaps layer outputs a tensor of size 10x16, which is used for 10-class classification.
2.2.3 Convolutional block attention module (CBAM)
The attention mechanism was initially applied to RNN models for image classification in 2014 [26]. Subsequently, in 2017, it was first introduced in the field of natural language processing [27]. In 2018, Woo et al. [28] proposed the Convolutional Block Attention Module (CBAM), which allows neural networks to focus more on information that plays a crucial role in the classification process while ignoring irrelevant information.
The attention mechanism in traditional convolutional networks mainly focuses on analyzing the channel domain, limiting the consideration of spatial relationships between feature map channels. In contrast, CBAM consists of two components: the channel attention module and the spatial attention module. By addressing both spatial and channel attention, CBAM overcomes the limitations of traditional attention mechanisms in capturing spatial relationship features between channels. The structure of CBAM is illustrated in Fig. 4.
The channel attention module starts by applying two parallel operations, MaxPool and AvgPool, to the input feature map, reducing its size to Channel\(\times \)1\(\times \)1. Then, the feature map is passed through a shared MLP module to compress the number of channels by a factor of n, followed by expansion through the ReLU activation function. The two output results are then added together. The output of the channel attention module is obtained by applying the sigmoid activation function. Finally, this output is multiplied element-wise with the original tensor, restoring it to the size of the original feature map. The process can be represented by Eq. (6).
The spatial attention module begins by applying MaxPool and AvgPool operations to the output results of the channel attention module, resulting in two feature maps. These two feature maps are then concatenated together. The concatenated feature map is passed through a 7\(\times \)7 convolutional layer, reducing it to a single-channel feature map. Finally, the sigmoid activation function is applied to obtain the feature map output of the spatial attention module. This output is multiplied element-wise with the original tensor, restoring it to the size of the original feature map. The process can be described by Eq. (7).
2.3 Proposed methodology
2.3.1 VGG-CapsNet
Inspired by the VGG network architecture and aiming to explore the impact of relative spatial positional information on Chest X-ray image classification, we propose a new network model called VGG-CapsNet. In VGG-CapsNet, we utilize convolutional layers with small-sized kernels from the VGG network to compress the images before inputting them into the capsule layers. The Capsule Network has shown excellent performance in simple images and has an irreplaceable advantage in recognizing overlapping images due to its adoption of the dynamic routing algorithm. However, the Capsule Network has significantly more parameters compared to traditional CNNs. For complex images with large pixel sizes, the number of parameters grows exponentially. Therefore, we use the 9\(\times \)9\(\times \)512 feature maps obtained from the feature extraction process using VGG19 as the input for the Capsule Network in our VGG-CapsNet model.
The VGG-CapsNet model takes an input size of 299\(\times \)299\(\times \)1 and produces an output size of 9\(\times \)9\(\times \)512 after passing through the VGG19 network. The 9\(\times \)9\(\times \)512 output is then reshaped and used as the input for the CapsNet. The primary capsule layer consists of 32 capsules, where each capsule is an 8-dimensional vector. The output of the primary capsule layer (3\(\times \)3\(\times \)32\(\times \)8) serves as the input for the digit capsule layer, which contains 2 capsules, each represented by a 16-dimensional vector. Each digit capsule receives input from (3\(\times \)3\(\times \)32) capsules, with each capsule being 8-dimensional. Using the routing-by-agreement algorithm, 2 16-dimensional vectors are computed and fed into a fully connected layer for binary classification, resulting in the probability of the input belonging to the COVID-19 class. The network structure of VGG-CapsNet, illustrated in Fig. 5, represents an example of a binary classification model.
2.3.2 CN2A-CapsNet
The VGG-CapsNet architecture addresses the issue of losing relative spatial positional information, but the choice of using VGG for feature extraction is often done blindly. Additionally, chest X-ray images exhibit redundancy and correlation properties, and it is not desirable to directly connect them to feature vectors. To overcome these challenges, we introduce an attention mechanism to dynamically determine weights and efficiently extract relevant features from X-ray images. To achieve this, we incorporate the channel attention and spatial attention modules from CBAM into the CNN architecture, resulting in the CN2A model. The CN2A model utilizes channel attention and spatial attention to adaptively adjust the importance of each channel and spatial position in the input and output feature maps. This allows it to effectively extract relevant features and leverage the capabilities of the CapsNet architecture [29,30,31,32].
In the CN2A-CapsNet architecture, there are two main components: CN2A and CapsNet. The CN2A part incorporates channel attention and spatial attention mechanisms to enhance the feature representation and select informative features. These refined features are then fed into the CapsNet component, which consists of primary capsules and digit capsules. The primary capsules generate vector outputs based on the adapted features, and the digit capsules further process these outputs for the final classification. The overall network structure of CN2A-CapsNet, illustrated in Table 1, demonstrates the integration of the CN2A module and the CapsNet architecture for the specific task of binary classification.
The CN2A-CapsNet network takes an input size of 299\(\times \)299\(\times \)1, and after feature extraction by the CN2A module, the output size becomes 9\(\times \)9\(\times \)512. This output is then reshaped and used as the input for the CapsNet component. The Primary Capsule layer in CapsNet consists of 32 capsules, with each capsule representing an 8-dimensional vector. The output of the Primary Capsule layer, which is of size 3\(\times \)3\(\times \)32\(\times \)8, is then passed to the Label Capsule layer. The Label Capsule layer contains 2 capsules, each with a carrier size of 16 dimensions. Each higher-level capsule receives input from 3\(\times \)3\(\times \)32 8-dimensional vectors. Using the dynamic routing algorithm, two 16-dimensional vectors are computed and passed to a fully connected layer to obtain the probability of a COVID-19-positive patient. The CN2A-CapsNet architecture consists of the CN2A module and the CapsNet component, and the schematic diagram of the network, exemplifying a binary classification model, is shown in Fig. 6.
3 Experiments and discussion
3.1 Performance indicators
In the assessment of classification outcomes, accuracy and recall are typically employed as measures. The calculation methods for these metrics are outlined in Table 2.
True Positive (TP) represents the number of cases correctly identified as COVID-19 positive. False Negative (FN) represents the number of cases erroneously identified as negative when they are positive. True Negative (TN) represents the number of cases correctly identified as negative. False Positive (FP) represents the number of cases erroneously identified as positive when they are negative.
As an auxiliary tool for medical diagnosis, we aim for this model to effectively identify positive cases. In this model, we choose accuracy and recall as evaluation metrics. The calculation methods for accuracy and recall are represented by Eqs. (8) and (9) respectively.
Accuracy reflects the overall performance of a model, while recall represents the proportion of correctly identified COVID-19 positive cases among all predicted positive cases. Recall holds high clinical significance, as a model with a high recall can serve as a machine-assisted tool in clinical practice for screening COVID-19-positive chest cases.
3.2 Cross-validation
To reflect the true performance of the model on the dataset and reduce errors, we employed cross-validation to randomly separate the training and testing sets. We used a 6-fold cross-validation method, dividing the dataset into six folds. In each iteration, five folds were used for training, while the remaining fold was used for testing. This process was repeated six times, rotating the folds each time. Finally, the results from the six iterations were averaged to obtain the final performance evaluation.
3.3 Simulation environment
The experimental network architecture was implemented using PyTorch. The simulation and testing were conducted on a personal computer running Ubuntu 20.04.4 LTS operating system. The computer was equipped with an Intel(R) Xeon(R) E5-2680 v4 CPU @ 2.40GHz \(\times \) 56 and an NVIDIA Corporation TU102 [GeForce RTX 2080 Ti] GPU.
For binary classification, we used the Stochastic Gradient Descent with Momentum (SGDM) optimizer with a learning rate of 0.001 and a momentum of 0.5. The batch size was set to 100. For the three-class classification, we used the Adam optimizer with a learning rate of 0.00005. The batch size was set to 200.
3.3.1 Experiment#1: impact of different input image sizes
In this experiment, our goal was to investigate the impact of different image sizes on classification results. By resizing the original dataset images (299\(\times \)299) to 224\(\times \)224 and 112\(\times \)112, we obtained three sets of images with different sizes. Subsequently, we conducted a three-class classification experiment using the VGG19 model on these three image sizes (299, 224, 112), and the experimental results are summarized in Table 3.
According to the data presented in the table, it can be observed that the time required to train one batch of images increases proportionally with the increase of input image size. However, further analysis of the experimental results indicates a significant improvement in classification accuracy as the input image size increases. In consideration of the trade-off between training time and classification accuracy, we prioritize the improvement of classification accuracy. Therefore, for the following experiments, we decided to use the original image size of the dataset (299\(\times \)299) as the input image size.
3.3.2 Experiment#2: VGG-CapsNet performance
In this experiment, we aimed to investigate the impact of relative spatial positional information in chest X-ray images on classification results. To offer an objective model evaluation, we conducted comprehensive experiments and analyses on both the VGG-CapsNet model and the VGG model. We evaluated the performance of the models using accuracy, recall, and training loss on the test set. Due to the varying complexity of binary and three-class classification tasks, we trained the models for 100 epochs in the binary classification task and 200 epochs in the three-class classification task. We present the results of one of the experiments for both binary and three-class classification tasks for the two models in Fig. 7.
Based on the experimental results, it can be observed that the VGG-CapsNet model outperforms the traditional VGG model in terms of both accuracy and recall on the test set. Particularly noteworthy is the significant advantage of VGG-CapsNet in terms of recall. These results indicate that the relative spatial positional information of chest X-ray images not only influences the classification results but also plays a crucial role in diagnosing COVID-19 patients. It’s worth noting that due to differences in data distribution within the public dataset, in our observations, we found that the VGG-CapsNet exhibited higher binary and ternary classification losses in the training set compared to the traditional VGG model. This suggests that the VGG model might have over-learned noise or irrelevant features, while VGG-CapsNet can better generalize the learned knowledge, demonstrating improved generalization ability. Therefore, through this experiment, we have discovered that capsule networks can effectively complement convolutional neural network structures, leading to improved network performance and better generalization ability.
Our research findings indicate that there is still potential for further improvement in feature extraction with VGG-CapsNet. To fully leverage the characteristics of capsule networks and input more efficient relevant information into the capsule network, we conducted Experiment 2 to validate the effectiveness of the CN2A model.
3.3.3 Experiment#3: CN2A-CapsNet performance
Although VGG-CapsNet is capable of effectively utilizing the relative spatial positional relationships between local features, we acknowledge that there is still room for improvement in its feature extraction. Our goal is to enhance the correlation between local features by introducing attention mechanisms and inputting more efficient features into the capsule network, thus fully leveraging the capabilities of the capsule network. To achieve this, we further propose the CN2A-CapsNet model by combining the capsule network with attention mechanisms.
This experiment aimed to investigate whether our proposed CN2A model could efficiently and accurately extract relevant information to further enhance network performance. To obtain an objective model evaluation, we conducted in-depth comparative experiments and analyses between CN2A-CapsNet and VGG-CapsNet. Accuracy, recall, and training set loss were used as performance metrics. In the experiment, we presented the results of one instance of each model for binary classification (trained for 100 epochs) and multi-class classification (trained for 200 epochs), as shown in Fig. 8.
According to the experimental results, the CN2A-CapsNet model, which incorporates the CN2A method, outperforms the VGG-CapsNet model. In the binary classification task, after 30 epochs of training, CN2A-CapsNet achieves stable accuracy and recall on the test set, gradually widening the gap with VGG-CapsNet. In the multi-class classification task, where the model needs to learn to differentiate more classes, after 100 epochs of training, CN2A-CapsNet starts to outperform VGG-CapsNet. By comparing the training set losses of the models, we observe that both models have similar losses, but CN2A-CapsNet exhibits higher accuracy. This indicates that CN2A-CapsNet better learns the key features in the training set, allowing it to classify new samples more accurately. In contrast, VGG-CapsNet overfits the noise in the training set, resulting in lower classification performance compared to CN2A-CapsNet. Therefore, in the chest X-ray image classification task of this experiment, our proposed CN2A model efficiently and accurately extracts relevant information and inputs more precise information into the capsule network, further enhancing the network performance.
To comprehensively evaluate the performance of CN2A-CapsNet, we employed a 6-fold cross-validation method. Firstly, the dataset of 2400 chest X-ray images was divided into 6 folds, with each fold containing 400 images. Then, in each iteration, 2000 images from 5 folds were used as the training set, while the remaining fold with 400 images was used as the test set. This process was repeated 6 times, ensuring that each fold served as the test set once. Finally, the results from the 6 iterations were summarized, and the average performance was presented in Fig. 9.
To provide a more intuitive evaluation of the models’ diagnostic performance, we represented the values in the confusion matrix as percentages. Figure 10 shows the confusion matrices generated for VGG-CapsNet and CN2A-CapsNet models for Fold-1. For the binary classification task, VGG-CapsNet exhibits a higher probability of correctly predicting COVID-19 patients (98.04%) compared to correctly predicting Normal cases (96.94%). Similarly, CN2A-CapsNet also demonstrates a higher probability of correctly predicting COVID-19 patients (99.02%) compared to correctly predicting Normal cases (98.98%). Therefore, CN2A-CapsNet possesses superior predictive performance for COVID-19 patients, with an error rate of 2% lower than VGG-CapsNet’s error rate of 5.02%. Regarding the three-class classification task, VGG-CapsNet shows a higher classification error rate for pneumonia cases, with 5.34% being incorrectly predicted. It is also prone to misclassifying COVID-19 patients and normal samples as pneumonia cases. Similarly, CN2A-CapsNet is influenced by pneumonia cases, with 3.82% of pneumonia cases being incorrectly predicted. However, it still leans towards accurate predictions for COVID-19 cases, which holds greater clinical significance in practical applications. The error rate of CN2A-CapsNet is 9.04%, lower than VGG-CapsNet’s error rate of 13.51%. Consequently, CN2A-CapsNet demonstrates a comparative advantage over VGG-CapsNet.
Based on the aforementioned experiments, we can draw the following conclusions: The results of Experiment 1 indicate that choosing the original image size of the dataset (299\(\times \)299) as the input size for the experiment was the most appropriate. The results of Experiment 2 indicate that the relative spatial positional information in chest X-ray images is an important factor in diagnosing COVID-19 patients. The results of Experiment 3 demonstrate that CN2A-CapsNet performs the best overall. The proposed CN2A model effectively extracts relevant information and feeds it into the capsule network, allowing the capsule network to fully utilize its characteristics and exhibit superior performance. However, due to variations in the methodologies and metrics used to evaluate each system, precise direct comparisons cannot be made, and these results are intended for reference purposes only.The strength of our proposed approach lies in its ability to achieve high recall rates without requiring an extensive amount of data samples for support.
3.3.4 Experiment#4: impact of different feature extractors
The aim of this experiment was to compare the performance of our proposed CN2A feature extractor with other feature extractors in a three-class classification task under identical dataset conditions. To ensure fairness, we utilized their respective source codes and conducted training on the same device. Table 4 presents the performance of the six feature extractors under identical conditions using the same classifier.
According to the experimental results, all six models utilizing CapsNet as the classifier exhibited higher recall rates than accuracy rates. This suggests that the relative spatial relationship information within chest X-ray images is crucial for diagnosing COVID-19 patients. Regarding the training time per batch, MobileNet V2, as a lightweight convolutional neural network model combined with the CapsNet classifier, demonstrated the shortest training time (10 s). However, it had the lowest accuracy among the six models. Inception V3, characterized by a mixed convolution and inception structure, had the longest training time per batch (47 s), yet its recall rate was lower than that of CN2A-CapsNet. The CN2A-CapsNet model outperformed the other five models in both accuracy and recall metrics. However, its training time was not the shortest. Nevertheless, considering all factors comprehensively, the CN2A-CapsNet model emerged as the optimal choice in terms of performance.
3.3.5 Proposed CN2A-CapsNet model performance comparison with baseline methods
We compared our proposed method with the approaches proposed by other researchers in previous studies, and the comparison results for binary classification and multi-class classification are presented in Tables 5 and 6, respectively. For the binary classification task, our method outperforms the methods used by Apostolopoulos et al. [33] and Hussain et al. [34] in terms of recall rate. Although their methods achieve slightly higher accuracy than ours, our recall rate significantly surpasses their results. In clinical practice, recall rate holds greater clinical significance. Compared to their methods, our model pays more attention to utilizing relative spatial positional features in chest X-ray images, which is crucial for COVID-19 diagnosis. Therefore, our method achieves much higher recall rate than theirs, despite a slight decrease in accuracy. Regarding Sharma et al. [35] and Hayat et al. [36], they did not disclose their recall rates. Although they use datasets that are significantly larger than ours, our method still performs better in terms of accuracy. For the three-class classification task, our method also has an advantage in terms of recall rate. Compared to the cited papers [33, 34], they used smaller datasets than ours, while our method surpasses them in both recall rate and accuracy, especially in recall rate. Compared to the methods used in the cited papers [35, 37, 38], our method still maintains an advantage in terms of accuracy and recall rate, even though they have larger datasets than ours. Compared to the CNN-LSTM model proposed by Islam et al. [39], their method outperforms our proposed method in both recall rate and accuracy. We believe this is mainly due to them employing data augmentation techniques on a portion of the dataset.
I have compared our proposed method with those of previous researchers on the same COVID-19 Radiography database (C19RD) and presented the comparison results in Table 7. Since most of the methods proposed by other researchers were only designed for a three-class classification problem, we also chose to compare under the same conditions. In comparison to the deep learning architecture F-RNN-LSTM proposed by Goyal et al. in reference [40], which utilizes a recursive neural network (RNN) with long short-term memory (LSTM), although their COVID-19 dataset had fewer samples than ours, the overall data scale was higher. However, our model achieved significantly higher accuracy and recall rates than the F-RNN-LSTM model. Compared to the CovidConvLSTM model proposed by Dey et al. [41], which encodes spatial correlations of the last CNN layer using convolutional long-short-term memory (ConvLSTM) layers, their accuracy is higher than our model, but their recall rate is 0.21% lower than ours. It should be noted that this accuracy difference may be due to the fact that their dataset size is six times larger than ours. In real-world clinical applications, recall rate and dataset size are critical factors for evaluating model performance. Compared to the method proposed by Kaya et al. [42], who selected five pre-trained models with the highest accuracy from fifteen CNN models and used them for feature extraction in D\(^{3}\)SENet, this strategy requires massive dataset support, and their accuracy and recall rate are based on the highest values in the training set. In contrast, our results are based on six-fold cross-validation averaging. Compared to the study in the cited paper [43], where the dataset size used in Fachrel J et al.’s CNN-LSTM model is four times larger than ours, their model performs 0.29% better than ours in terms of accuracy on the basis of a large amount of training data. However, in terms of recall rate, our model has a significant advantage, surpassing their model by 2.34%. Additionally, compared to the Multi-head+Self-attention model proposed by Ukwuoma CC in the cited paper [44], their dataset size is five times larger than ours. Nevertheless, our model outperforms theirs in both accuracy and recall rate.
Overall, we have observed that previous research methods tend to achieve higher accuracy than recall rate, whereas our model achieves a higher recall rate than accuracy. This is because our study focuses on the clinical significance of recall rate when applied in practice. However, in the field of deep learning, accuracy and recall rate are often in conflict, and balancing between them is an important indicator for evaluating model performance. Our proposed method has a significant advantage in achieving high recall rate without requiring a large number of data samples. This advantage lies in our utilization of the understanding ability of capsule network for spatial information, in which the captured relevant features are further processed by the capsule network. In addition, we have proposed the CN2A model, which efficiently mines relevant information from chest X-ray images by enhancing the focus of convolutional neural networks on important regions. Ultimately, we have successfully attained high recall rates on a limited dataset. However, due to variations in the methodologies and metrics used to evaluate each system, precise direct comparisons cannot be made, and these results are intended for reference purposes only. The strength of our proposed approach lies in its ability to achieve high recall rates without requiring an extensive amount of data samples for support.
4 Discussion
Experiment 1 established that using the original image size of the dataset (299\(\times \)299) as the input size was the most suitable choice. Subsequently, experiment 2 demonstrated that integrating the Capsule Network with the traditional CNN model resulted in improvements in accuracy by 0.75% and recall by 1.69% for binary classification tasks. For three-class classification tasks, accuracy increased by 2.75% and recall by 2.51%. Please refer to Fig. 9 for detailed results. These findings suggest that capsule networks effectively complement traditional CNN models. The limitations of traditional CNN models in maintaining relationships between object parts and the overall context are evident, particularly due to the loss of relative spatial relationship information among elements caused by max pooling operations. In contrast, capsule networks leverage vector representations to capture the relative spatial positional features within images, leading to enhanced model performance. Furthermore, the experimental results further validate the critical role of relative spatial positional relationships in diagnosing COVID-19 patients using chest X-ray images. This underscores the paramount importance of leveraging relative spatial positional features for the diagnosis of COVID-19 patients.
Experiment 3 demonstrates a significant performance improvement achieved by our proposed CN2A model, which combines traditional CNN architecture with an attention mechanism. In binary classification tasks, the CN2A model exhibited a 1.25% increase in accuracy and a 1.26% improvement in recall. For ternary classification tasks, the model showcased an accuracy enhancement of 1.67% and a recall improvement of 1.92%. These results indicate that the CN2A model effectively integrates CBAM with traditional CNN models. This successful integration is attributed to the inclusion of the Channel Attention Module and Spatial Attention Module in the CBAM model, which addresses the limitations of traditional attention mechanisms in capturing inter-channel spatial relationship features. The Channel Attention Module in the CBAM model aligns precisely with the required inter-channel spatial relationship features. Additionally, the introduction of the attention mechanism in the CN2A model enhances focus on important regions, further refining the extraction of critical features from chest X-ray images. Furthermore, it is evident that the CN2A model exhibits higher improvements in accuracy and recall rates for ternary classification tasks compared to binary classification tasks. This indicates that the CN2A model more effectively exploits the pertinent information crucial for classification, particularly demonstrating its advantages in complex classification scenarios. Experiment 4 further validated the superiority of integrating the CN2A feature extractor with the CapsNet classifier by comparing its performance with five other feature extractors on a three-class classification task.
5 Conclusions
This study proposes a deep learning model called CN2A-CapsNet for diagnosing COVID-19 cases from chest X-ray images. Compared to methods proposed by other researchers, our model achieves a high recall rate on a small-scale dataset, making it more suitable for real-world applications. Specifically, in experiment one, we determined that using the original image size of the dataset (299\(\times \)299) as the input size was the most appropriate choice. Experiment two compared the results of VGG-CapsNet and VGG19 models, validating the effectiveness of the Capsule Network in complementing the convolutional neural network structure. We also discovered the significance of relative spatial relationships in chest X-ray images for diagnosing COVID-19 patients. Subsequently, in experiment three, we compared the results of the CN2A-CapsNet and VGG-CapsNet models, demonstrating that our proposed CN2A model efficiently captures relevant information, leading to further improvements in network performance. Finally, our experiments in Experiment Four revealed that all six models utilizing CapsNet as the classifier exhibited higher recall rates than accuracy rates. This finding suggests that employing CapsNet as the classifier emphasizes the utilization of relative spatial positional relationships in chest X-ray images, which is crucial for diagnosing COVID-19 patients. Moreover, from a comprehensive perspective, the CN2A-CapsNet model emerged as the optimal choice in terms of performance. Moreover, we compared our CN2A-CapsNet model with other methods proposed by researchers, demonstrating significant advantages, namely achieving high recall rates with limited data samples.
In summary, CN2A-CapsNet, through its efficient feature extraction method, achieves a high recall rate on a small-scale dataset, providing a practical solution for controlling the spread of COVID-19. However, in practical applications, chest X-ray images of patients may suffer from inaccuracies such as motion artifacts, foreign object shadows, and overexposure. Therefore, future work will focus on researching and addressing these issues to fully leverage the limited chest X-ray images of patients and achieve accurate diagnoses.
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: https://www.kaggle.com/datasets /tawsifurrahman/covid19-radiography-database (accessed on March 11th, 2023).
References
Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (covid-19): a review. JAMA. 2020;324(8):782–93.
Xu T, Shao M, Liu R, et al. Risk perception, perceived government coping validity, and individual response in the early stage of the covid-19 pandemic in china. Int J Environ Res Public Health. 2023;20(3):1982.
Alsagheir A, Amer S, Alzubaidi L, et al. A national survey of children, adults, and the elderly in the fourth wave of the Covid-19 pandemic to compare acute and post-Covid-19 conditions in Saudi Arabia. J Clin Med. 2023;12(6):2242.
weekly epidemiological update C. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—1-september-2023. WHO. Accessed 16 Oct 2023
Dugdale CM, Walensky RP. Filtration efficiency, effectiveness, and availability of n95 face masks for covid-19 prevention. JAMA Intern Med. 2020;180(12):1612–3.
Zhou T, Liu F, Lu H, et al. A review of deep learning imaging diagnostic methods for covid-19. Electronics. 2023;12(5):1167.
Ukwuoma CC, Qin Z, Heyat MBB, et al. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest x-ray images. J Adv Res. 2023;48:191–211.
Luo JX, Liu WF, Yu L. Pneumothorax recognition neural network based on feature fusion of frontal and lateral chest x-ray images. IEEE Access. 2022;10:53175–87.
Shan F, Gao Y, Wang J, et al (2020) Lung infection quantification of covid-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655
Dey N, Zhang YD, Rajinikanth V, et al. Customized vgg19 architecture for pneumonia detection in chest x-rays. Pattern Recogn Lett. 2021;143:67–74.
Awassa L, Jdey I, Dhahri H, et al. Study of different deep learning methods for coronavirus (covid-19) pandemic: taxonomy, survey and insights. Sensors. 2022;22(5):1890.
Alahmari SS, Altazi B, Hwang J, et al. A comprehensive review of deep learning-based methods for covid-19 detection using chest x-ray images. IEEE Access. 2022. https://doi.org/10.1109/ACCESS.2022.3208138.
Mohammad-Rahimi H, Nadimi M, Ghalyanchi-Langeroudi A, et al. Application of machine learning in diagnosis of covid-19 through x-ray and CT images: a scoping review. Front Cardiovasc Med. 2021;8:638011.
Alshmrani GMM, Ni Q, Jiang R, et al. A deep learning architecture for multi-class lung diseases classification using chest x-ray (CXR) images. Alex Eng J. 2023;64:923–35.
Roy S, Tyagi M, Bansal V, et al. Svd-clahe boosting and balanced loss function for covid-19 detection from an imbalanced chest x-ray dataset. Comput Biol Med. 2022;150: 106092.
Er MB. Covid-19 detection based on pre-trained deep networks and LSTM model using x-ray images enhanced contrast with artificial bee colony algorithm. Expert Syst. 2023;40(3):e13185.
Lee CP, Lim KM. Covid-19 diagnosis on chest radiographs with enhanced deep neural networks. Diagnostics. 2022;12(8):1828.
Kaya Y, Gürsoy E. A mobilenet-based cnn model with a novel fine-tuning mechanism for covid-19 infection detection. Soft Comput. 2023;27(9):5521–35.
Avazov K, Abdusalomov A, Mukhiddinov M, et al. An improvement for the automatic classification method for ultrasound images used on CNN. Int J Wavelets Multiresolut Inf Process. 2022;20(02):2150054.
Nahiduzzaman M, Goni MOF, Islam MR, et al. Detection of various lung diseases including covid-19 using extreme learning machine algorithm based on the features extracted from a lightweight CNN architecture. Biocybern Biomed Eng. 2023. https://doi.org/10.1016/j.bbe.2023.06.003.
Sabour S, Frosst N, Hinton GE (2017) Dynamic routing between capsules. In: Advances in neural information processing systems 30
AbouEl-Magd LM, Darwish A, Snasel V, et al. A pre-trained convolutional neural network with optimized capsule networks for chest x-rays covid-19 diagnosis. Clust Comput. 2023;26(2):1389–403.
Chowdhury ME, Rahman T, Khandakar A, et al. Can AI help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–76.
Rahman T, Khandakar A, Qiblawey Y, et al. Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Comput Biol Med. 2021;132:104319.
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Mnih V, Heess N, Graves A (2014) Recurrent models of visual attention. In: Advances in Neural Information Processing Systems 27
Vaswani A, Shazeer N, Parmar N, et al (2017) Attention is all you need. In: Advances in neural information processing systems 30
Woo S, Park J, Lee JY, et al (2018) Cbam: convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Xu Z, Wang Y, Xiong W, et al. A novel attentional feature fusion with inception based on capsule network and application to the fault diagnosis of bearing with small data samples. Machines. 2022;10(9):789.
Xu Z, Lee CKM, Lv Y, et al. Ensemble capsule network with an attention mechanism for the fault diagnosis of bearings from imbalanced data samples. Sensors. 2022;22(15):5543.
Li P, Han T, Ren Y, et al. Improved yolov4-tiny based on attention mechanism for skin detection. PeerJ Comput Sci. 2023;9:e1288.
Qi L, Lu X, Shen H, et al. Automatic classification of mass shape and margin on mammography with artificial intelligence: deep CNN versus radiomics. J Digit Imaging. 2023. https://doi.org/10.1007/s10278-023-00798-w.
Aim TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635.
Hussain E, Hasan M, Rahman MA, et al. Corodet: a deep learning based classification for covid-19 detection using chest x-ray images. Chaos Solitons Fractals. 2021;142:110495.
Sharma P, Arya R, Verma R, et al. Conv-CapsNet: capsule based network for covid-19 detection through x-ray scans. Multimed Tools Appl. 2023. https://doi.org/10.1007/s11042-023-14353-w.
Ahatsham H, Baglat P, Mendonça F, et al. Novel comparative study for the detection of covid-19 using CT scan and chest x-ray images. Int J Environ Res Public Health. 2023;20(2):1268.
Sharma A, Singh K, Koundal D. A novel fusion based convolutional neural network approach for classification of covid-19 from chest x-ray images. Biomed Signal Process Control. 2022;77:103778.
Chakraborty M, Dhavale SV, Ingole J. Corona-Nidaan: lightweight deep convolutional neural network for chest x-ray based covid-19 infection detection. Appl Intell. 2021;51(5):3026–43.
Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (covid-19) using x-ray images. Inform Med Unlocked. 2020;20:100412.
Goyal S, Singh R. Detection and classification of lung diseases for pneumonia and covid-19 using machine and deep learning techniques. J Ambient Intell Humaniz Comput. 2021. https://doi.org/10.1007/s12652-021-03464-7.
Dey S, Bhattacharya R, Malakar S, et al. Covidconvlstm: a fuzzy ensemble model for covid-19 detection from chest x-rays. Expert Syst Appl. 2022;206:117812.
Kaya M, Eris M. D3senet: a hybrid deep feature extraction network for covid-19 classification using chest x-ray images. Biomed Signal Process Control. 2023;82:104559.
Fachrel J, Pravitasari AA, Yulita IN, et al. Enhancing an imbalanced lung disease x-ray image classification with the CNN-LSTM model. Appl Sci. 2023;13(14):8227.
Ukwuoma CC, Cai D, Heyat MBB, et al. Deep learning framework for rapid and accurate respiratory covid-19 prediction using chest x-ray images. J King Saud Univ-Comput Inf Sci. 2023;35(7):101596.
Funding
This study was funded by the Scientific Research Foundation of the Education Department of Heilongjiang Province under grant 145209409.
Author information
Authors and Affiliations
Contributions
Conceptualization: HZ, ZL Methodology: HZ, ZL, SL Data Collection: ZL, SL, ZS Manuscript writing: ZL, ZZ Writing original draft preparation: ZL, ZZ Resources: HZ, ZZ Supervision: HZ, ZS
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article has been published with the written informed consent of all authors.
Competing interests
The authors have not disclosed any Conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, H., Lv, Z., Liu, S. et al. Cn2a-capsnet: a capsule network and CNN-attention based method for COVID-19 chest X-ray image diagnosis. Discov Appl Sci 6, 190 (2024). https://doi.org/10.1007/s42452-024-05796-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-024-05796-3