Abstract
Cyberphysical microelectrode-dot-array (CP-MEDA)-based digital microfluidic biochip (DMFB) is attracting more attention than its predecessor of traditional DMFB. Conventional DMFBs are mostly unable to recover from the errors incurred at assay run time, and thus, it leads to unacceptable results. Recent studies have revealed the vulnerability of CP-DFMBs to detect malicious intrusions during its design or operational phase. In this paper, we have analysed such vulnerable scenarios that have been utilized by the routing-based-synthesis approach on a CP-MEDA. We have also depicted the mixing operations based on routing for ensuring better security measures. Various attacking scenarios have been demonstrated on a MEDA-based DMFB, and moreover, checkpoint-based intrusion detection method has been proposed for the RBS technique. The effectiveness of our approach is compared with the available benchmark assays. Our proposed method has shown significant improvement over the existing state-of-the-art procedures in terms of assay execution time and intrusion detection rate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In the recent past, microfluidic biochips are extensively used for on-chip implementation of several in vitro bio-protocols or laboratory assays, usually needed in medical diagnostics [37]. These lab-on-a-chip (LoC) devices have introduced a paradigm shift in DNA analysis, toxicity grading, molecular biology, drug design, automated drug delivery, and threat assessment against bio-terrorism [35]. It essentially offers a viable and low-cost alternative for reducing health care costs of cardiovascular diseases, cancer, diabetes, for providing point-of-care (P-O-C) health services [36]. These DMFB chips are expected to be immensely useful for rapid and accurate diagnosis of various diseases including malaria, HIV virus, etc., and for mitigating neglected tropical diseases prevalent in developing countries [31]. DMFB can simplify cumbersome laboratory procedures by manipulating fluids at nanolitre (\(10^{-9}\)) or picolitre (\(10^{-12}\)) volume scale [32] with minimal human intervention. Thus, it leads to high throughput, sensitivity, and accuracy of test results compared to traditional benchmark procedures [18, 20].
In recent years, MEDA-based DMFB architecture has been proposed in [24, 41, 42]. Each MEDA-cell consists of a group of micro-electrodes or a sea-of-microelectrodes and an activation circuit [13]. Similar to a dot-matrix printer, the dynamic grouping of microelectrodes have been done to form various shapes and sizes of the droplets on a MEDA-based DMFB. It can be activated simultaneously to perform microfluidic operations by controlling the associated activation circuits. Sensing response time for MEDA (10 ms) is attractively high relative to the conventional DMFB [43] (30 s) due to the combination of active CMOS logic integrated circuits with each microelectrode [13].
Integration of cyberphysical paradigm to DMFBs can be analysed in the various phases of a DMFB design flow. The flow can be compromised by an attacker that leads to undesirable consequences [4]. CAD tools are utilized to convert a high-level assay specification into an actuation sequence that accomplishes the biochip synthesis [38, 39]. The intrusion of malicious components (Trojans) in the foundries is quite common [22]. During fabrication procedure, intellectual property (IP)-based CAD tools are usually procured from different third-party IP (3PIP) vendors. Malicious components having such a characteristic are commonly termed as Hardware Trojan Horses (HTH).
In this work, we have proposed a MEDA-based routing-based-synthesis (RBS) method and a routing path alteration procedure in case of a malicious attack on RBS. In the RBS method, the entire synthesis is accomplished without using any dedicated mixing modules on the chip. All mixing operations of a bio-protocol are performed based on different shift patterns [8]. We have found out the mixing completion time using Lagrange’s interpolation formula [8] and also proposed a new shift pattern for MEDA biochips. The results on the benchmark data sets depict that the proposed method has significantly reduced the overall synthesis time and space consumption on the chip. However, the efficiency of routing has been increased by many folds in this method, as all mixing operations are based on diffusion model [33]. Also, several attacking scenarios are demonstrated for entire method on a MEDA-based DMFB. The effectiveness of our technique is established by comparing the results with available benchmark assays. We have shown much faster assay execution rate compared to module-based synthesis by adopting the proposed RBS methodology. It is also shown that the checkpoint-based intrusion(error) detection rate is quite significant based on the proposed technique.
The key contributions of our paper are as follows:
-
i)
In this work, we have proposed a novel approach for the bio-synthesis procedure that would reduce the bioassay completion time and ensure better security.
-
ii)
A novel \(45^{0}\)-shift movement (diagonal movement) approach only for the MEDA-based biochips has been discussed. Also, its mixing capability for the RBS method has been incorporated.
-
iii)
We have also proposed an approach for analysing checkpoint-based error detection and recovery mechanisms to mitigate such attacks.
The remaining manuscript is structured as follows: In Sect. 2, we have discussed the motivation behind the project. In Sect. 3, the preliminary concepts of MEDA biochips basic construction, droplet manipulations, synthesis steps, and threat model for DMFB have been discussed. The enhanced routing-based synthesis (RBS) mechanism is presented for MEDA biochips in Sect. 4, where we have to compute the time steps required for each directional shift and their corresponding mixing completion percentage. Two different types of mixing modules are also presented in this section. In Sect. 5, a new kind of \(8\times 8\) assay architecture is given for PCR bioassay chip along with illustrative examples for the proposed method. Finally, the synthesis process using RBS and checkpoint-based security approach for RBS is given in Sect. 6. Section 7 represents the simulation results in detail and finally, in Sect. 8, we have concluded.
2 Motivation
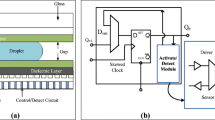
Our primary motivation behind the MEDA-based biochip is due to its flexibility compared to the conventional DMFB. Fluidic procedures like routing, mixing, splitting, merging, and detection on MEDA-based biochips can be finished in considerably less time compared to conventional DMFBs. Droplet routing time is very much relevant which is not considered in earlier works [29, 44]. A droplet can move in any direction, and highly integrated microelectrode cell (MC) acts as sensors for detecting an error that occurs during the synthesis procedure. A fixed module is placed on the array, and the entire mixing is done over that fixed module. Maximum cells remain unused at the time of mixing. The security implications of MEDA for the implementation of experimental setups and laboratory schedules on remotely accessed robotic systems are the other important motivations for research. In recent years, a few works have been developed in remotely control laboratories for the implementation of biochemistry protocols [1, 2]. Cyber protection remains a major concern to date as these protocols are downloaded to remote servers. Over the next few years, these are projected that automated laboratories will be miniaturized to MEDA-based lab-on-chip. Security issues for such laboratory automation systems are therefore also important to emerging MEDA. A high-level synthesis flow diagram for MEDA biochips [25] is shown in Fig. 1 where a unified priority controller, scheduler, placer, and the router are entreated. Reservoir locations are primarily allocated using reservoir placer.
Synthesis flow for MEDA-based biochips [25]
MEDA presents the slanting development of droplets that gives a more prominent level of opportunity contrasted with customary DMFBs where bead development is restricted to the “horizontal” and “vertical” movements. This level of opportunity can be misused for more productive bioassay execution. Due to the incorporation of dynamic CMOS rationale, detecting can be done anyplace on a MEDA-based biochip [4], and the reaction time for detecting (10ms [5]) is a lot more modest contrasted with that required on a regular DMFB (e.g. the 30s, as revealed in [7]). The sensors must be coordinated in explicit territories of the biochip that makes the present DMFBs disadvantageous. MEDA permits far greater adaptability as a sensor can be coordinated beneath each electrode.
The bioassay synthesis on a MEDA biochip is performed through the co-optimize operations like module binding, scheduling [19], placement, and routing [39] as shown in Fig. 2. Complete design flows for MEDA biochips have been made possible with help of extensive research. As a result, the biological assay can be represented in a high-level languages termed as biocoder [5]. Indeed, automated protocol synthesis and realizations for MEDA biochips design performed a significant level of sophistication in recent times using architectural and physical synthesis tools [38, 39] which are being widely used across the biological-Loc industry. An automated horizontal custom MEDA biochips design flow is shown in Fig. 2.
3 Preliminaries
3.1 Microelectrode dot array-based biochips architecture
In a MEDA biochip, various kinds of biomedical sample droplets in micro-/nano-volume have lied on a series of controllable micro-electrodes. Blood, serum, urine, and saliva are mainly taken as sample droplets which are sandwiched in between two parallel glass plates as shown in Fig. 3. Silicone oil is used as a filler medium. The bottom plate consists of highly integrated micro-electrodes, and the top plate of the entire chip is used as a ground electrode. The droplet will therefore be moved horizontally, vertically, or diagonally using the principle of ‘electrowetting-on-dielectric’ (EWOD) [16]. As compared with conventional DMFBs, MEDA biochips have the real-time capacitive sensing property called “droplet-property sensing” and trace the location of the droplet using the “droplet-location sensing” property [46].
3.1.1 MEDA architecture
MEDA biochips are comprised of several regular EWOD microfluidic components called dot array microelectrode. Compared to the standard biochips, MEDA \(\approx\) 9 to 16 times smaller unit microelectrode cells than those used by normal DMFB [13]. Advanced MEDA architecture allows efficient reconfiguration and has also carried out various forms of fluidic operation on the chip, such as sorting, blending, slicing, and dilution [9].
Droplet size depends upon the number of microelectrodes on which a droplet is resting. To indicate the position of the droplet (in the sea of micro-electrodes) [27], one reference point R is fixed at the corner microelectrode on which droplet is lying, red colour point as shown in Fig. 4a. H is the height and W is the width of the droplet. B is called bounding width, which signifies the minimum distance required to separate two droplets as shown in Fig. 4. A droplet can dynamically change its droplet aspect ratio during routing [27] by grouping multiple microelectrodes so that functional electrodes are formed. A typical microdroplet with \(4\times 4\) shape is shown in Fig. 4a, and four droplets of different sizes \(2\times 2\), \(6\times 4\), \(4\times 4\) and \(6\times 6\) are occupying 4, 24, 16, and 36 micro-electrodes, respectively, are depicted in Fig. 4b. We have mainly considered the typical microdroplet with \(4\times 4\) shape for our entire synthesis process. The droplet that lies on the sixteen microelectrodes and thirty-two microelectrodes are required to route from one cell to another cell like conventional DMFB.
3.2 Various kind of routing constraints
In the proposed RBS method, contamination-free routing is our prime concern because the entire mixing (dilution) is performed using droplet routing. One single droplet can lie on series of micro-dot arrays. If we have assumed that initially, the size of all droplets is the same. If it can utilize a single cell in DMFB, it is equivalent to 9 or 16 cells in MEDA [15]. Two types of fluidic constraints are maintained for fault-free routing: One is static and the other is the dynamic fluidic constraint.
Minimum microelectrode gap required at any time instant ‘t’ between two droplets \(D^{t}_{i}\) and \(D^{t}_{j}\) is called static fluidic constraints, where \(j\in i\) but \(j\ne i\). The width of two mixer droplets \(D^{t}_{i}(x,y)\) and \(D^{t}_{j}(x,y)\) is \(W_{j}\) and \(W_{i}\), respectively. We have assumed that the width of the droplets is same, i.e. \(W_{i}=W_{j}\)=W. To satisfy the static constraint which is given by the equation, two droplets should maintain the gap.
Similarly, during dynamic movements of the droplets at the very next time instant (t+1), the minimum microelectrode gap required between two is named dynamic fluidic constraints. To satisfy dynamic fluidic constraints, minimum space required between two droplets \(D^{t}_{i}\) and \(D^{t}_{j}\) with coordinate values \((X^{t}_{i}\),\(Y^{t}_{i})\) and \((X^{t}_{j}\),\(Y^{t}_{j})\), respectively, at time instant t or next (t+1) has to be \(\ge\) (W+1).
3.3 Threat model for CP DMFB
To improve the performance of the biochips and give the scope of efficient error-recovery for the biochips, cyberphysical inclusion attracts various possible attacks to the DMF chip at the same time. The malicious biocoder/designer manipulates the result of the assay consequence and separates the assay conditions [3]. Further, it will modify the concentration of the sample, incubation time, and mixing time. Various kinds of such attacks are listed in Table 1. Examples of such attacks in cyberphysical system like stealth attacks, replay attacks and covert attacks [6] are the example of static attack [30]. They lead to output attacks that are consistent with the measurements equation, system dynamics, and reset the measurements and measurements can be cancelled, respectively. If the placement is finished, the routing algorithm decides the optimum path of the individual test operation droplets according to the schedule constraints. It also takes into account fluidic restrictions, such as the microelectrodes gap between two droplets, to avoid unintended mixing of two or more than two droplets. The output of the droplet routing step is the actuation sequence that stores the droplet movement control information at each step. False data injection attacks are dynamic considered as a special type of output attack rendering an unstable mode (if any) of the system unobservant [34]. Apart from the above-mentioned attacks, some other attacks [3] explicitly can happen to a CP-DMFB due to the following reasons:
-
(a)
Malicious Bio-coder/Designer of the DMFB
-
(b)
Malicious CAD tool vendor (third parties)
-
(c)
Denial of Service attack due to hardware Trojans
-
(d)
Attacks on test results manipulation
-
(e)
Attacks on Control Software
-
(f)
Violating the minimum spacing between droplets
-
(g)
Malicious modification of droplet route
3.4 Completion time
The experimental results were carried out by Paik et al. [33] to show the time needed for different mixing modules. Laboratory tests on different mixing modules were conducted in [33], and a module library has been prepared for conventional DMFB, which is available in Table 2.
4 Problem formulation
We are considering the PCR bioassay [23] and a \(8\times 9\) traditional module-based biochip as shown in Fig. 5 for problem formulation. To accomplished first layer of PCR, 4 mixing modules (\(M_{1}\), \(M_{2}\), \(M_{3}\), and \(M_{4}\)) of two \(1\times 4\), \(2\times 3\) and \(2\times 4\) mixer-unit, respectively, had been placed on the DMFB chip. Two mixing operations \(M_{1}\), \(M_{2}\) are performed on \(1\times 4\) mixer-units simultaneously and also \(M_{3}\), \(M_{4}\) have run on \(1\times 4\) and \(1\times 3\) mixer units, respectively. Then, \(M_{1}\), \(M_{2}\) will be completed in 4.6 sec (the time required for 100% of \(1\times 4\) modules shown in Table 2), but \(M_{3}\), \(M_{4}\) operations will take 2.9 sec. and 6.1 sec, respectively. The next layer of PCR \(M_{5}\) is the mixing of \(M_{1}\) and \(M_{2}\); thus, \(M_{5}\) can directly start after 4.6 sec, whereas \(M_{6}\) can begin after the completion of \(M_{4}\) (6.1 sec.). Now if someone has to consider \(1\times 4\) and \(2\times 4\) mixer-unit (faster mixing units present on the chip) for \(M_{5}\) and \(M_{6}\), respectively, the minimum time required for mixing completion is 7.5 sec. and 10.6 sec, respectively.
It is also evident that M7 can only be done after \(13.6 \approx 14\)sec. The completion time of the PCR assay thus requires 14 secs in module-based DMFB ignoring the time needed for other operations such as separating, merging, and droplet identification, in addition, to match the first layer of the PCR sequence graph (G) with 4 parallel mixing operations (nodes) as applicable. No other way will the modules be mounted on a \(8 \times 9\) (\(72 \times 16\) microelectrodes \(\equiv 1152\) microelectrodes, if we consider one cell used for conventional DMFB is equivalent to 16 microelectrodes for MEDA) chip size over the module sizes to provide more free cells for other operations. Of the 72 cells (1152 microelectrodes) in all, there are only 9 cells (144 microelectrodes) required for other operations such as identification, dispensing, etc. Therefore, the module-based approach to synthesis required more synthesis time and suffered more operating costs in case of an error.
4.1 Routing path alteration attack on RBS process
The possible chances of an attack on the routing path via alteration of routing of a previously scheduled droplet [12] (actuation sequences) increased numerously. There could be various reasons for which routing path alteration attack occurs.
CASE 1, During the fabrication process Trojans en-grafted on biochips
Modification of high-level assay specifications/sequence graph (G) and low-level electrode actuation sequences can lead to denial-of-service (DoS) attacks. We focus our attention purely on the MEDA biochip itself, and on the attacks that can be considered a form of hardware Trojan [12] on RBS process.
CASE 2, Trojans embedded in Third-Party Vendor’s Routing files
Intrusion can be made through the control software, and point-of-care embedded systems are physically vulnerable to modification. As a result, the actuation sequence of the biochips can be modified and that will bring other malicious droplets into the RBS mixing path. We have primarily considered routing file modification attack, which leads to actuation sequence alteration in the present work.
5 Proposed approach of mixing
An efficient routing-based synthesis (RBS) process has been proposed to ensure less time required for the bioassay completion compared to the existing methods. The technique also ensures the adaptation of better security measures on the chip based on the checkpoints.
There are two types of modules present in MBS method which is represented as \(1 \times N\) (linear array of cell) and \(2 \times N\) (two-dimensional array) as a display in Figs. 6 and 7, respectively, where \(N \in \mathbb {Z^{+}}\) and the values of N lies between 2 to infinity. To determine the entire synthesis completion time, we have to find out the mixing completion percentage of each shifting movement. Suppose the frequency (f) of the entire system is chosen 16 Hz as mention in [33]. As we know, f is inversely proportional to time. Then, the time (t) required to move \(i^{th}\) droplet \(D^{i}\) from its current coordinate position \(D^{i}_{x,y}\) to any of one of its adjacent eight electrodes is 0.0625 sec, where \(i \in {\mathbb {Z}}^{+}\) and (x,y) is the coordinate position. There are three types of shift movement present in general in a traditional DMFB. Type-I \(0^{0}\), Type-II \(90^{0}\), and Type-III \(180^{0}\) are generally termed as zero-degree, ninety-degree, and one hundred eighty-degree shift movements, respectively. Now the \(0^{0}\) movements can be categorized into two types. \(0^{0}_{1}\) and \(0^{0}_{2}\) (one zero-degree drift and two consecutive linear zero-degrees drift, respectively) present in \(1\times 3\), \(2\times 3\) and \(1\times 4\) and \(2\times 4\) MBS framework, respectively.
Only \(180^{0}\) shift movement can be accomplished 100% mixing in \(1 \times 2\) modules [33]. The entire mixing completion time (frequency of the system taken as 16 Hz [33]) for \(1 \times 2\) module is equivalent to 17 sec. So the number of shift required for 100% mixing is \(\frac{17}{0.0625}\) = 272 steps. Similarly, the number of \(90^{0}\) shifts required for \(2 \times 2\) modules (Total mixing time = 9.95 sec. [29, 33]) is \(\frac{9.95}{0.0625}\) = \(\lceil 159.2 \rceil\) \(\cong\) 160 steps (round up the value with the pessimistic assumption). It has been shown that due to flow reversibility present in \(1 \times N\) frame work [29], we have chosen the next bigger module \(2 \times N\) framework for RBS method. In the presence of multiple pivot points \(2 \times N\), it will accelerate the entire mixing process and it also takes a lesser mixing completion time as compared to \(1 \times N\) framework.
Now we can compute the mixing percentage for a single step of \(90^{0}\) shift. With the help of 160 steps \(90^{0}\) shift movement can accomplish 100% of mixing. One single step of \(90^{0}\) shift can accomplish is equal to \(\frac{100}{160}\) \(\cong\) 0.625% of mixing which is shown in Table 3. From Table 2, the time required to accomplish 100% mixing in \(2 \times 3\) and \(2 \times 4\) is 6.1 and 2.9 sec, respectively. In these two mixing modules, entire mixing operation has been done with the help of \(90^{0}\) and \(0^{0}\) shift movements.
Now, we have to consider \(2\times N\) mixing modules as shown in Fig. 7 to calculate the mixing percentage of a single or multiple \(0^{0}\)-shifts. Figure 7b represents a \(2\times 3\) module in which one \(0^{0}\)-shift is followed by two consecutive \(90^{0}\)-shift or vice versa repeated simultaneously until the completion of 100% mixing. Total numbers of time steps required for \(2\times 3\) module are \(\frac{6}{0.0625}\) equal to 96. Hence, one single \(0^{0}\)-shift can accomplished \(\frac{((100-(64\times 0.625)))}{32}\) = 1.875%. Similarly from \(2\times 4\) mixing module, we can compute \((0^{0}_{2})\) (consecutive two zero degree shift) which is equal to \(\approx\) 7%.
Similarly, the percentage of completion mixing for different linear shift motions \((0^{0}_{1}\) and \(0^{0}_{2})\) and \(90^{0}\)-shifts is also determined from the \(2\times N\) mixing system and the results are shown in Table 3.
5.1 Proposed mixing architecture
It has been seen that more numbers of a straight run (consecutive \(0^{0}\)-shifts) can accelerate the mixing completion time or rather say increase the percentage of mixing on traditional as well as MEDA biochips. If we have compared between two existing mixing modules \(1\times 4\) and \(2\times 2\), both are acquired an equal number of cells but the time required to accomplished 100% of mixing \(1\times 4\) module can take lesser time than that of \(2\times 2\) module. It entrenched the fact that to increase the mixing percentage different shift patterns are responsible. Hence, overall synthesis time for MBS method [33] decreases if mixing in done only on mixing module present in \(2\times N_{max.}\) framework, where \(N_{max.}\) is fixed at 4. This is also a drawback for MBS method.
To remove such kind of limitation in MBS, \(2 \times N\) framework is being considered for our RBS method, where \(N_{max.} \ge 4\). Extending the value of N is very much desire for RBS method. Hence to find out the completion time required for the entire dilution, first we have to compute \(0^{0}_{3}\), \(0^{0}_{4}\),....,\(0^{0}_{n}\), where \(n \ge 2\) and \(n \in {\mathbb {Z}}\).
Our intention is to achieve more numbers of linear shifts; with this regards, we have to find out the next bigger mixing patterns present in \(2 \times N\) framework. To minimize the mixing completion time, we have required such kind of shift pattern, where N must be greater than 4. Lagrange’s interpolation formula [8] is applied to compute the probable completion time required for mixing (dilution) in the very next bigger mixing \(2 \times N\) framework. Now we have considered \(x_{0}\), \(x_{1}\), \(x_{2}\), \(x_{3},..., x_{n}\) as the values of N on \(2 \times N\) framework and \(y_{0}\), \(y_{1}\), \(y_{2}\), \(y_{3},..., y_{n}\) denotes mixing completion time. Hence, the layout is shaped using Lagrange’s interpolation formula as follows:
From Eq. 4 it has been seen that the values of L(x) are parabolic polynomial in nature. By taking the first derivative of L(x), i.e. \(L^{\prime }(x)\)=(x-6.5), we can see that minima exit -6.5, and the value of vertex is present in -0.125. According to that time required for \(2\times 6\), \(2\times 7\) mixing modules (where 4 and 5 straight runs present) for entire dilution is zero. For practical experiments this is an infeasible situation.
Using the curve fitting method, we have fitted the curve by simulating 1000 times (fewer data points available) on a curve-fitting simulator and a more negative estimation of mixing time in different modules was considered to suit the curve correctly. Repeated tests provided a more realistic calculation. We may use the trend contained in the curve fitted values and observe the underlying error characteristics using the Gaussian distribution. It is also easy to say that the probability density for the Gaussian distribution [7] to find the true values around the actual is as follows:
Equation 5 represents the probability that a given measurement will have a value x. The probability that the measurement falls between x and x + dx is given as P(x)dx. Since measurement will have some value on the real number, it follows that \(1=\int _{-\infty }^{\infty }P(x)dx\). There are a number of observations that can be made to find 5; the function which is symmetric with respect to \(\mu = x\). It decreases more rapidly with smaller \(\sigma\). The standard deviation is a measure of when we report the average value of N measurements, the uncertainty we should associate with this average value is the standard deviation of the mean \(\mu\), often called the standard error \(\pm \sigma\). Now \(\sigma = \sqrt{(x-\mu )^{2}}\), error observed by a single experiment. Hence, the error % is \(\sigma \le 0.02\) which is less significant and we derive the L(x) as follows:
It has been observed from Equation No. 6 that the mixing completion time increases till N = 6 and it repetition shows after \(N\ge 7\) as shown in Fig. 9. We have therefore proposed a novel mixer architecture as \(2\times N\) framework. The mixing patterns are given in Fig. 8 where a droplet may run three or four consecutive linear shift movements for mixing.
On MEDA-based biochips, each droplet has some flexibility to move \(45^{0}\) or \(60^{0}\) in the bioassay. So, we have to find out the mixing percentage of completion for that particular movement. We have already derived the mixing completion of \(180^{0}\) and \(90^{0}\). Using these two values, we can calculate the percentage of mixing for \(45^{0}\) or \(60^{0}\) movements as given below.
Putting the value of x as 45 and 60 for \(45^{0}\) and \(60^{0}\) movements, respectively, we have got the value of 0.745% and 0.7%, respectively, which are more than the \(180^{0}\) and \(90^{0}\) moves.
It has been observed that \(N\ge 7\) amounts, the achievement of more repetitive motions (patterns) does not increase the mixing speed. The execution period is quite degraded as seen in Fig. 9. Therefore, after 4 consecutive linear shifts \((0^{0}_{4}\), the necessary turbulence is added in our shift pattern in the form of a mandatory \(45^{0}\), \(60^{0}\) and \(90^{0}\) change which definitely improves the mixing time. In Table 3, the respective mixing completion percentage for \(45^{0}\), \(0^{0}_{3}\) and \(0^{0}_{4}\) is also provided in violet colour. Total of mixing finish after 1, 2 or more \(0^{0}\)-shifts are seen as \(SR_{sum}(n)=\sum ^{n}_{i=1}0^{0}_{i}\), where \(i \in \mathbb {Z^{+}}\) and \(SR_{sum}(n)\) is denoted as Straight Run mixing percentage after consecutive \(n^{th}\) \(0^{0}\) shift. Thus, we can increase the shift movement by increasing the value of n and find the mixing completion for a consecutive straight run which are as follows:
Here \(0^{0}_{1}\) denoted as single \(0^{0}\)-shift from one cell to another cell, similarly \(0^{0}_{2}\) signifies two zero degree move. In the same manner we have represented \(0^{0}_{3}\), \(0^{0}_{4}\), ..., \(0^{0}_{i}\).
The proposed shift patterns in the RBS mixing system used these shift patterns to different precedence order. They were built-in descending order according to their percentage of mixing as \({0}^{0}_{4}> 0^{0}_{3}> 0^{0}_{2}> 0^{0}_{1}> 45^{0}> 90^{0} > 180^{0}\).
According to RBS, if a \(0^{0}_{1}\)-shift is open, then the droplet appears to achieve more consecutive straight-run movements \(0^{0}_{2}\), \(0^{0}_{3}\) or \(0^{0}_{4}\)-shift in a greedy way before a collision occurs with other droplets or exhausted path (checking the assay’s boundary wall) for more linear movement. Where both the left and right distances are equal as described in the pseudo-code shown in Fig.10, depending on the cell coordinate location. The droplet moved up and right, co-parenting its child [8] in the next sequencing graph step.
Sequential graph (G), routing constraint (\(\delta\)), maximum array size (A) and source–target of each net have been initialized in the first phase of the Algorithm 1 shown in Fig. 10. Time steps after each movement are automatically updated, and it also checks the error (in terms of intrusion) after \(16^{th}\)-time steps. If there is any error find, it will automatically initiate the roll-forward module and go to the initial level. Hence, the proposed algorithm (Fig. 10) chooses the priority movement from the set of shift_movements (\(\eta\)). The droplet can take four consecutive \(0^{0}\)-shift from its current position obeying routing constraint (\(\delta\)). It will take a necessary \(45^{0}\) move after completion of four consecutive \(0^{0}\) shift movements. After successful completion of 100 percent mixing, it will call Mix_Set algorithm for splitting and merging of two droplets according to the sequential graph (G).
-
The new mixer droplet formation algorithm is given in Fig. 1, in which tearing the mixer droplet retains the routing protocol (\(\delta\)) and concerns about the assay margin (\(\lambda\)) (boundary wall of the assay through which it is properly split out). Primary goal of our newly proposed Algorithm 2 is to protect accidental mixing between any random droplets. In the previous Algorithm [10], routing constraints and boundary wall concepts were not considered.
-
Completely mixed droplets are divided into two identical droplets and prepared to be combined with another reagent or sample droplets according to sequence graph (G). Hence, we have computed the co-coordinates (x,y) of all the co-parents of the mixer droplets. Minimum Euclidean distance (ED) between all the co-parents is selected as the target microelectrodes, and the corresponding droplets are redirected to that target electrodes as shown in Fig. 1. In conventional DMFB [10], minimum distance is found with the basis of Manhattan distance.
-
Merging of two mixer droplets at the very next step of the scheduling graph is based on the minimum Euclidean distance between them. Effective completion of the merger of two droplets would initiate Algorithm 1 if it is not in the last stage of the scheduling graph. The mixing was started again for the next synthesis stage according to the sequence graph (G), and the process continues until the entire synthesis process would completed.
-
Detection unit was not present in the previous algorithm [10]. After successful completion of the scheduling graph, the mixer droplet must be tested using a detection module that is incorporated in the current version of Algorithm 2.
In RBS, we have attempted to adopt the most efficient mixing trends of a module of \(2\times 6\). The droplet has to take a necessary \(45^{0}\) or \(90^{0}\)-shift for every \(0^{0}_{4}\)-shift. In the following example, the minimum time steps (t) needed in RBS for mixing completion are given.
Example If we have assumed 16 Hz operating frequency of the chip, the minimum number of time-steps required for full mixing completion (100%) is 27t where t= 1 time-step.
As per the proposed RBS method, the maximum amount of mixing completion can be achieved by the \(0^{0}_{1-4}\)-shift (18.875%), which consists of four consecutive straight runs of the mixer droplet. After having a \(0^{0}_{4}\)-shift, it is mandatory to take an \(45^{0}\) or \(90^{0}\)-move before acquiring another linear movement (\(0^{0}_{4}\), \(0^{0}_{3}\), \(0^{0}_{2}\) or at least a \(0^{0}_{1}\)). Now, from Table 3 it is seen that one \(0^{0}_{4}\) along with one \(90^{0}-move\) can accomplish \(0^{0}_{4}\) + \(90^{0}\) \(\Rightarrow\) (18.875 + 0.625)% = 19.5% of mixing completion. Hence, remaining mixing to be accomplished = (100-19.5)% = 80.5%.
Again from Table 3, it is evident that the fastest possible remaining mixing can be completed if the remaining mixing pattern consists of \(0^{0}_{4}\) and \(90^{0}\)-shift only for conventional DMFB as well as \(0^{0}_{4}\) and \(45^{0}\)-shift for MEDA biochips alternately one followed by the other as shown in Fig 11a and b. If such a shift pattern can be set for the remaining mixing on the chip (without considering the routing congestion among different mixing paths), then total mixing completion in \(1^{st}\) cycle is computed for DMFB and MEDA, respectively. In Fig. 11a and b, \(0^{0}_{4}\), \(90^{0}\)-shifts and \(0^{0}_{4}\), \(45^{0}\)-shifts are, respectively, represented by blue and red colour in \(1^{st}\) cycle and by green and red arrow for the \(2^{nd}\) mixing cycle.
The mixing completion for DMFB and MEDA computed for the first cycle and that is \((0^{0}_{4} + 90^{0}+ 0^{0}_{4} + 90^{0}+ 0^{0}_{4} == 78 \%\) and \(90^{0}+ 0^{0}_{4} + 90^{0})=78.48\%\), respectively. The time required for 78% or 78.48% of mixing is 4*(4t) + 4*(t) =20t, where t = 1 time-step. As we can see that due to \(45^{0}\)-shift, MEDA mixing percentage is slightly greater than the conventional DMFB. The total time or movement required to accomplish the 100% mixing of a mixer droplet is 27t steps as shown in Fig. 11a and b.
We can also choose \(0^{0}_{3}\)-shift or \(0^{0}_{2}\) or \(0^{0}_{1}\)-shift in our shift pattern. To get the scenario, we have to delete the \(0^{0}_{4}\)-shift (the \(4^{th}\) linear shift) from Fig. 11a and b. In place of that we must need to insert one \(45^{0}\) or \(90^{0}\)-shift and then again we may search for \(0^{0}_{4}\)-shift if available. In any case the present \(4^{th}\) shift in the sub-cycle which is now (\(45^{0}\) or \(90^{0}\)) always accomplish lesser amount of mixing (Mix_percentage) compared to \(0^{0}_{4}\) alone. Hence, in any case, minimum steps required will be 27t and the optimum shift patterns are shown in Fig. 11a and b.
6 Synthesis process using RBS
In RBS method, we have proposed different shift pattern based on which mixing operations are accomplished. The percentage of mixing completion for each shift-movement is shown in Table 3. Also in RBS method precedence of choosing various shift-movement is as follows:
\(O^{0}_{4}\)-shift > \(O^{0}_{3}\)-shift > \(O^{0}_{2}\)-shift > \(O^{0}_{1}\)-shift > \(45^{0}\)-shift > \(90^{0}\)-shift > \(180^{0}\)-shift.
In RBS process a new shift pattern (\(45^{0}\)-shift) is incorporated only for MEDA biochips, unlike conventional DMFB. In the present work, the RBS method is used to incorporate checkpoint-based intrusion detection effectively.
6.1 RBS-chip architecture
In Fig. 12, a standard \(8\times 8\) Application Specific Integrated Circuit (ASIC) chip is pictured for example. Here more than four mixing operations numbers (\(> 4\)) can be performed simultaneously in larger chip sizes.
From the patterns of the derived shifts shown in Fig. 11a and b, we have proposed unique RBS chip architecture [10] (for \(M_{j} \le 4;\) for any layer \(L_{i} \exists G\)). The re-configurable operations (mixing) are carried out along the chip’s boundary cells, as shown in Fig. 13a. For traditional DMFB, Fig. 13b is seen. The MEDA biochips are depicted in Fig. 13c and d. The chip’s centre region is left out for non-re-configurable operations, such as containing droplet reservoir detection units (\(D_{i}\)), heater and error recovery. Obviously, such an architecture eliminates the costs of routing between different stages of a bio-protocol. It would allow for optimum operation of the RBS synthesis with a symmetric pattern obtained for all mixing operations \(M_{1}\), ..., \(M_{4}\) for both DMFB and MEDA-based biochips.
6.2 Various mixing layers for PCR with the given example
Different PCR stages for mixing shown in Fig. 5 consist of a number of \(M_{j}\) operations, where j is denoted from one to seven mixing operations. In the first layer of PCR, mixer droplet route by RBS method for simultaneous mixing operations. \(M_{1}\), \(M_{2}\), \(M_{3}\) and \(M_{4}\) mixing steps represented by red, green, violet, and blue colours, respectively, which is shown in Fig. 13a, b, c and d for a \(8\times 8\) cells DMFB and \(8\times 8 \times 16\) microelectrodes-based MEDA biochip, respectively.
A mixer droplet route according to our proposed shift patterns from 1st to 17th and 18th to 31st time-step, respectively, as shown in Fig. 13a and b shows path. Similarly in Fig. 13c and d the corresponding mixing completion percentages are computed for MEDA \(1^{st}\) to \(14^{th}\) and \(15^{th}\) to \(27^{st}\) time-step, respectively. Complete mixing completion time and corresponding mixing completion percentage up to \(31^{st}\) and \(27^{th}\) time-steps for traditional DMFB as well as MEDA are given in Table 4 and 5.
The example in Fig. 12 shows the primary arrangement for a PCR assay, and Fig. 13a–d represents the respective layer 1 mixing patterns that are symmetrical to each other. For DMFB the time-steps(t) shifts obtained are given in Equation No. 7 and 8.
Table 4 shows the rate of mixing in % with respect to time for each shift pattern of a PCR assay. Hence, from Fig. 13c and d it has been clear that a lesser amount of time required to accomplish 100% mixing completion for MEDA-based biochips compared to traditional DMFB is computed in Table 5. Shift patterns with escaped time-steps(t) for MEDA are given in Equation No. 9 and 10.
RBS pattern on highly integrated MEDA-based biochips also decreases the overall cost for washing. The cells which are common between two or more heterogeneous fluid [9] need to be washed on conventional DMFB. Hence, the corresponding mixer droplets (after 100% mixing completion) and their respective coordinate positions in subscript form after \(28^{th}\) time steps are depicted on Fig. 13b which are \(M_{1(7,5)}\), \(M_{2(4,7)}\), \(M_{3(5,2)}\), and \(M_{4(6,8)}\). Before starting very next layer of PCR mixing operations, we need to split \(M_{1}\), \(M_{2}\), \(M_{3}\) and \(M_{4}\) mixer droplets and merge accordingly using the Split_and_Merge algorithm (algorithm is shown in Fig. 14) which will require another 4 time-stamps as shown in Fig. 15a–b.
From the sequence graph of PCR, another two mixing operations \(M_{5}\) and \(M_{6}\) can be started on \(31^{st}\) time stamp as shown in Fig. 15c and it is accomplished at \(57^{th}\) time stamp. Similarly, completion of 100% mixing \(M_{5}\) and \(M_{6}\) needs to split and merge accordingly to form \(M_{7}\). Final layer of PCR, \(M_{7}\) will be started at \(60^{th}\) time steps shown in Fig. 16a. \(M_{7}\) needs another 27 time steps to accomplished 100% mixing as pictured in Fig. 16b. Entire PCR assay operation completed the mixing at \(87^{th}\) time-stamp.
The time needed to accomplished the entire PCR assay synthesis is \((87 \times 0.0625) = 5.4375\)sec. About 55 percent improvement in accordance with DMFB based earlier module approaches exists. Similarly, our proposed method tested on exiting benchmark assays [11] as well as some hard research benches [11], and the results obtained are quite impressive.
6.2.1 Routing route modification attack on RBS cycle
In RBS, the frequency of the routing increases several times due to all of the diffusion-based mixing operations [33]. It is done by detecting different shift patterns on the chip. Also growing are potential chances of routing route modification attack (RRMA).
CASE 1: For consecutive 4 time steps a mixer droplet will take a straight run (\(0^{0}\)-Shift). After that, there has to be a compulsory \(45^{0}\) or \(90^{0}\)-shift. It happens that the droplet runs straight for more than three consecutive \(0^{0} {4}\)) due to actuation sequence modification, as shown in Fig. A 17a. We have called a scenario of an attack like “\(0^{0}\)-shift overrun attack”.
CASE 2: For droplet ’M j’ (Fig. 17b), while space is available for a straight run but M j has been unnecessarily redirected earlier and has taken a \(45^{0}\) or \(90^{0}\)-shift. The actuation sequence has been altered in the middle of \(0^{0}\)-shift, and this connection is considered for RBS system as “direct run violation attack.”
6.2.2 Checkpoint-based attack detection
A malicious droplet detection system based on a checkpoint is proposed to solve the security problems on RBS. MEDA integrated sensors will track the complete progress of an assay. The sensors are contrasted with the [40] assay execution reference behaviour. If biochip can be tracked for the whole duration of an assay execution at-time stage (t), we may detect 100 percent intrusion. Such a scheme is time- and overhead costs computationally intensive, and inefficient. Implementation is not feasible. The proposed checkpoint arrangements developed into RBS method are as follows:
-
Check points for Form 1: In RBS, the \(M_{1}\), \(M_{2}\), \(M_{3}\) and \(M_{4}\) mixing operations are started from the chip’s cell position coordinates (2,2), (7,2), (2,7) and (7,7), respectively. The first type of (static) checkpoint is placed on the starting cell from which each mixing process is initiated. These static checkpoints will be disabled until the 100% percent completion of the mixing is achieved. It will make sure the dispenser will detect if any other malicious droplet reaches the entire assay for PCR layer 1 within that time.
-
Check points for Form 2: The second form of the static checkpoint is put to test whether for a mixer droplet to have a \(0^{0}_{5}\)-shift taken. It has the provision to push \(0^{0}_{5}\)-shift more straight. By Fig. 17 it is possible to see that at (x , y) = (6, 1) coordinate chip location. A checkpoint will detect whether or not any alteration occurred. Because of that, the droplet \(M_{1}\) (BLUE) mistakenly moves to (6,1) location that is not needed according to our proposed algorithm.
-
Check points for Form 3: The third dynamic checkpoint is designated at 3rd and 4th time steps of each mixing operations. It will detect that the droplet can take unnecessary \(45^{0}\) or \(90^{0}\) instead of \(0^{0}\)-shift. It will also position the droplet provision of possibly \(45^{0}\) or \(90^{0}\)-shift instead of \(180^{0}\) inside the mixing change movements at \(5^{th}\) time steps. Desired facilities for putting all forms of checkpoints are displayed in Fig. 17.
6.3 Error recovery strategy
Various error recovery strategies are tabulated for standard DMFB operations as shown in Table 6. We have proposed a recovery mechanism in case of any error happened and which were undetected by the checkpoints as explained in the last section. In RBS approach, suppose the operating frequency of the chip is 16 Hz. Then, 1 time-step (t) = (1/16) = 0.0625 sec. The recovery problem is formulated to find the optimum time step (\(t_{d}\)) at which integrated sensor-based monitoring should be placed so that the recovery overhead (in terms of the traversed path) will be minimum. In RBS shift patterns after completion of entire mixing if an error found, a repeat of the entire operation. To determine mixing completion, we need to wait until the end of the mixing operation. To tackle the RBS recovery problem, the following assumptions are taken into consideration.
Assumptions
-
In RBS approach, the average time required to accomplish 100% mixing is 32t, i.e. the average path to be traversed by a mixer-droplet is fixed to be 32 to complete the mixing.
-
The entire path (\(n_{0}\), \(n_{1}\), \(n_{2}\),..., \(n_{31}\)) consists of all 32 distinct cells (each cell is equivalent to 16 micro electrodes) on the chip traversed from \(t_{0}\) time step to \(t_{31}\), i.e. no cell is repeated twice on the path (P) as shown in Fig. 18 and no STALL operation is required in these 32 steps.
-
High sensitivity MEDA-based bio-chips are assumed to be a ‘fair-chip’. Hence, the probability of each microelectrode cell on the entire path length (P = \(32 \times 16\)) being defected which belongs to that path, i.e. probability of each cell being defected on the path is 1/P.
Based on the above assumptions, physical-aware software can be integrated with RBS method. It can read and analyse sensor data and dynamically adapt the given synthesis for cyberphysical chips. Hence, our technique can be updated with a modified sequence graph, scheduling operations, and droplet routing pathways in run-time. Unlike [21], RBS control software does not need to consider module placement and resource binding phase. It minimizes initial synthesis time as well as online resynthesis time after monitoring the assay. Here, we do not target monitoring the chip for every time-steps (t) of the entire synthesis duration, which is expensive and computationally intensive for the control software.
The chip has to be monitored only at a single intermediate time instant (\(t_{d}\)) during the entire operation for once and after completion of the final mixing operation. The output will be monitored ( at \(t_{f}\)) for once only. Thus, incorporating an extra intermediate checkpoint will minimize the re-synthesis overhead at runtime and makes the entire error recovery much faster for RBS approach. At time-step \(t_{d}\), if we monitor the droplet, then we can easily roll back the operation from \(t_{d}\).
For example, if we find the error as early as at \(8^{th}\) time step, i.e. \(t_{d} = 8\) and \(1 \ge t_{d} \ge 32\). We shall not continue the operation any more till the end (\(t_{f}\)) and roll back from \(t_{d} = 8^{th}\) time-step itself. If we assume the next run to be fault free and the said operation finishes successfully at \(t_{f} = 32t\) in next run, then the total time required to finish the operation would be: \(T_{d} + T_{f} = 8 + 32 = 40t\) Now, according to Assumption No. II of Section 6.3, the number of faults which can be detected at intermediate checkpoint (\(t_{d}\)) would be 8 and the remaining 24 faults may happen at (cell number \(n_{9}\) to cell number \(n_{31}\)) cannot be detected at intermediate checkpoint (\(t_{d}\)). It may only be detected at final checkpoint (\(t_{f}\)). Because of equally likely probability of having an individual cell being defected on the entire path length (Assumption No. III of Section 6.3), the total recovery time (\(T_{R}\)) required would be as follows for large number of trials.
With the above example if \(T_{d} = 8\) and \(T_{f} = 32\) ; total time-steps (\(T_{R}\)) required recovering from the errors would be as follows:
Similarly, if we fix the intermediate detection at \(T_{d}\) = 10 and \(T_{f}\) = 32
and for \(T_{d}\) = 12 and \(T_{f}\) = 32
Putting different numerical values in \(T_{d}\), we have got the parabolic curve where \(T_{R}\) initially decreases and then again increase as shown in Fig. 19. If the early intermediate detection checkpoint is fixed at \(T_{d}\) = 16 time step, i.e. exactly at the halfway path, the overall recovery time can be minimized. So, we have fixed the detection twice, first at \(t_{d}\) = 16t after the mixing operation gets started and finally at completion (\(T_{f}\) = 32t), i.e. after finishing of the entire operation.
Lemma 1
The error recovery time will be minimized for RBS method if the intermediate error detection checkpoint is fixed at n= N/2 time-steps, where N is the total path length.
Proof
To generalize the above concept we have assumed:
-
N = total path length;
-
n = intermediate single checkpoint
-
P = total path travelled to complete a mixing operation including rollback
Now we have simulated the same mixing operation for a very large number of trials. According to the theory of classical probability, ’n’ number of faults may happen at first ’n’ cells on the path and can be detected at \(T_{d}\) Fig. 19. The remaining (N-n) number of faults will happen on rest of the path length and can be detected at final checkpoint (\(T_{f}\)) only. For first ‘n’ number of faults on the RBS-path the distance traversed by the droplet is:
For remaining (N-n) faults the total distance to be traversed by the droplet is:
So the total path (P) calculated from Equation No. 12 and 13 :
Equation No. 14 is a symmetric function and we need to find the value of ‘n’ for which the total recovery path as well as operation completion path (S) should be minimum. Thus, finding derivative from Equation No. 14 is as follows:-
Putting, (\(\frac{dp}{dn}\)) = 0; we find n = N/2; and (\(\frac{d^{2}p}{d^{2}n}\)) = 2, which is a positive integer and sufficient condition for existing minima at n = N/2. Hence, the above claim of n= N/2 is proved. Hence, the intermediate detection (checkpoint) is fixed exactly at halfway path of the proposed RBS-shift patterns. \(\square\)
6.4 Example
If at the time stage n=16 a certain mixing operation fails to achieve the necessary threshold values (threshold set according to the biochemical assay sequence graph and biochemistry nature), then that particular mixing operation is rolling back without interrupting others. Thus, RBS method does not test for a mixing completion threshold value at higher time steps and does not wait for rollback until the entire mixing is complete, unlike the module-based synthesis. Mistakes are detected here much more easily and rollback activity can be initiated earlier (at t=16). The unused droplets are stored at the previous level, and can be used for faster rollback before the next mixing stage is complete. Such deposited droplets are discarded into the waste reservoir until the next stage of mixing reaches completion of 100 percent. This cycle continues until the full synthesis of the bioassay is full.
7 Experimental results
The prime motto of RBS algorithm is to reduce overall synthesis time as well as complete the entire synthesis process without any kind of error intrusion. Simulations result of our proposed RBS algorithm in cyberphysical MEDA biochips have been reduced entire synthesis time significantly for real the life bioassay like PCR, In_vitro, CPA, IDP [45] and PDNA [28]. The entire simulation process has been performed on an Intel(R) core(TM) i3-8130U CPU with 4GB installed memory (RAM) and 64-bit operating system.
7.1 Real life assays tests
The RBS synthesis technique is being studied both on traditional DMFB and MEDA biochips. This is therefore compared to other module-based synthesis techniques, such as ILP and Tabu search [29] on physiological fluid IVD and PCR benchmarks. The test is performed for different chip dimensions and sizes (48 cells to 144 cells is equivalent to \(48\times 16\) to \(144\times 16\) microelectrodes, respectively ) and the synthesis time of the bioassay is promisingly improved by about 45% as shown in Table 7.
7.2 Comparative study between chip size and completion time
The proposed RBS has also been run for PCR using various chip size from \(8\times 8\) to \(16\times 16\) (49 to 256 cells or \(49\times 16\) to \(256\times 16\) microelectrodes), and the completion varies with size of the chip as shown in Table 8. We have tested 4 PCR (stage I) mixing operations simultaneously on various chip sizes. Completion-time decreases when four parallel mixing operations has been performed on the chip size of \(12\times 12\). After that increment of chip size does not take place in the completion period. So we may infer that a maximum of \(12\times 12\) chip size is adequate for four simultaneous mixing. It will also achieve minimum time by RBS patterns. From Table 9, it is very important to note that the synthesis time improvement up to 35% for real-life assays is measured on a \(12\times 12\) chip.
7.3 Analysis of mixing completion time for BS I & BS III as well as some hard benchmarks
Our proposed RBS algorithm for MEDA has been simulated on some of the existing benchmarks. We have also tested RBS algorithm with conventional as well as MEDA biochips on 6 randomly taken harder test cases [11]. It has been taken from the hard test benchmark which are commonly termed as Benchmark Suite III (BS-III) [14]. BS-III have 30 numbers of test cases. Our prime target to reduce synthesis time for the parallel mixing operations where the number is \(\ge 8\) in a single layer of sequence graph. Naturally, it will increases the overall assay synthesis time and also error recovery time. It ultimately increases the complexity and number of sensors requirement. RBS on MEDA-based biochips performed 100% synthesis for entire test cases from BS-I and it decreases the synthesis time below 7 sec. shown in Tables 10 and 11 which is quite promising. The average overhead time taken for RBS is well below 8% which is also quite promising.
7.4 Error detection rate on RBS
Some major problems on recovery operations are associated with SMG-based [26] and PTA-based [17] methods. Splitting error can be rectified by splitting re-execution, moving process, and reuse of droplets which has been stored in the previous level. However, MEDA-specific fluidic operations (e.g. droplet aliquot [46]) are not completely utilized for error recovery. The static and dynamic checkpoints can detect most of the errors (\(\ge 90\%\)) for the given example problem and PCR bio-assay as shown in Table 12. Comparative study between MEDA-based exiting method with our proposed method is given on Table 12. We have observed an average detection rate well above 90% that is quite satisfactory by placing of static and dynamic checkpoints on RBS.
8 Conclusion
The proposed MEDA-based routing synthesis method has accelerated assay execution by eradicating the format idea about dedicated implicit modules presence on the traditional DMFB. The entire mixing operation can be performed using the droplet routing according to the proposed shift movements (patterns) through the available path (microelectrode-dot-array) present on the MEDA biochips. Also integration of checkpoint-based error detection technique and the method for minimization of error recovery overhead would allow implementation of more high-performance bio-assays. Assay synthesis time has been reduced significantly in our proposed routing-based synthesis according to the experimental results as well as error (intrusion) detection rates. Washing is required if more numbers of heterogeneous droplets sharing the same path on the chip that be incorporated in the future for more accurate results. Also, we are planning for examining the pattern of static checkpoints and checkpoint optimization as the extension of this work.
References
Emeraldcloudlab. (2016) The emerald cloud laboratory [online]. Available: http://www.emeraldcloudlab.com
Transcriptic. (2016) Automated cell and molecular biology laboratory [online]. Available: https://www.transcriptic.com
Ali SS, Ibrahim M, Sinanoglu O, Chakrabarty K, Karri R (2015) Security assessment of cyberphysical digital microfluidic biochips. IEEE/ACM Trans Comput Biol Bioinform 13(3):445–458
Ali SS, Ibrahim M, Sinanoglu O, Chakrabarty K, Karri R (2016) Microfluidic encryption of on-chip biochemical assays. In: 2016 IEEE Biomedical Circuits and Systems Conference (BioCAS), pp 152–155
Ananthanarayanan Vaishnavi, Thies W (2010) Biocoder: a programming language for standardizing and automating biology protocols. J Biol Eng. https://doi.org/10.1186/1754-1611-4-13
Basu S, Saha S, Pan I (2014) Intrusion detection in online controller of digital microfluidic biochips. In: 2014 International Conference on Computational Intelligence and Communication Networks, IEEE, pp 1021–1025
Cartwright S (2003) Introduction to experimental error https://www.sheffield.ac.uk/polopoly_fs/1.14221!/file/IntroToExperimentalErrors_y2
Chakraborty S, Chakraborty S (2017) A novel approach towards biochemical synthesis on cyberphysical digital microfluidic biochip. In: VLSI Design and 2017 16th International Conference on Embedded Systems (VLSID), 2017 30th International Conference on, IEEE, pp 355–360
Chakraborty S, Chakraborty S (2019) Routing performance optimization for homogeneous droplets on meda-based digital microfluidic biochips. In: 2019 IEEE Computer Society Annual Symposium on VLSI (ISVLSI), IEEE, pp 419–424
Chakraborty S, Chakraborty S (2020) An efficient module-less synthesis approach for digital microfluidic biochip. SN Appl Sci 2(8):1–18
Chakraborty S, Chakraborty S, Das C, Dasgupta P (2016) Efficient two phase heuristic routing technique for digital microfluidic biochip. IET Comput Dig Tech 10(5):233–242
Chakraborty S, Das C, Chakraborty S (2018) Securing module-less synthesis on cyberphysical digital microfluidic biochips from malicious intrusions. In: 2018 31st International Conference on VLSI Design and 2018 17th International Conference on Embedded Systems (VLSID), IEEE, pp 467–468
Chen Z, Teng DHY, Wang GCJ, Fan SK (2011) Droplet routing in high-level synthesis of configurable digital microfluidic biochips based on microelectrode dot array architecture. BioChip J 5(4):343–352. https://doi.org/10.1007/s13206-011-5408-5
Cho M, Pan DZ (2008) A high-performance droplet routing algorithm for digital microfluidic biochips. IEEE Trans Comput-Aid Design Int Circ Syst 27(10):1714–1724
Chung W, Cheng P, Li Z, Ho T (2018) Module placement under completion-time uncertainty in micro-electrode-dot-array digital microfluidic biochips. IEEE Trans Multi-Scale Comput Syst 4(4):811–821. https://doi.org/10.1109/TMSCS.2018.2822799
Ding J, Chakrabarty K, Fair RB (2001) Scheduling of microfluidic operations for reconfigurable two-dimensional electrowetting arrays. IEEE Trans Comput-Aid Design Int Circ Syst 20(12):1463–1468. https://doi.org/10.1109/43.969439
Elfar M, Zhong Z, Li Z, Chakrabarty K, Pajic M (2017) Synthesis of error-recovery protocols for micro-electrode-dot-array digital microfluidic biochips. ACM Trans Embedd Comput Syst (TECS) 16(5s):1–22
Fair RB, Khlystov A, Tailor TD, Ivanov V, Evans RD, Srinivasan V, Pamula VK, Pollack MG, Griffin PB, Zhou J (2007) Chemical and biological applications of digital-microfluidic devices. IEEE Design Test Comput 24(1):10–24. https://doi.org/10.1109/MDT.2007.8
Grissom D, Brisk P (2012) Path scheduling on digital microfluidic biochips. DAC Design Automation Conference 2012:26–35. https://doi.org/10.1145/2228360.2228367
Ho TY, Chakrabarty K, Pop P (2011) Digital microfluidic biochips: recent research and emerging challenges. In: Proceedings of the seventh IEEE/ACM/IFIP International Conference on Hardware/software codesign and system synthesis, ACM, pp 335–344
Ibrahim M, Chakrabarty K (2015) Efficient error recovery in cyberphysical digital-microfluidic biochips. IEEE Trans Multi-Scale Comput Syst 1(1):46–58
Karri R, Rajendran J, Rosenfeld K, Tehranipoor M (2010) Trustworthy hardware: identifying and classifying hardware trojans. Computer 43(10):39–46. https://doi.org/10.1109/MC.2010.299
Kramer MF, Coen DM (2001) Enzymatic amplification of dna by pcr: standard procedures and optimization. Curr protocol Mol Biol 56(1):1–15
Lai KYT, Yang YT, Lee CY (2015) An intelligent digital microfluidic processor for biomedical detection. J Sig Process Syst 78(1):85–93. https://doi.org/10.1007/s11265-014-0939-3
Li Z, Ho T, Lai KY, Chakrabarty K, Yu P, Lee C (2016) High-level synthesis for micro-electrode-dot-array digital microfluidic biochips. In: 2016 53nd ACM/EDAC/IEEE Design Automation Conference (DAC), pp 1–6
Li Z, Lai KYT, McCrone J, Yu PH, Chakrabarty K, Pajic M, Ho TY, Lee CY (2017a) Efficient and adaptive error recovery in a micro-electrode-dot-array digital microfluidic biochip. IEEE Trans Comput-Aid Design Inte Circ Syst 37(3):601–614
Li Z, Lai KYT, Yu PH, Chakrabarty K, Ho TY, Lee CY (2017b) Droplet size-aware high-level synthesis for micro-electrode-dot-array digital microfluidic biochips. IEEE Trans Biomed Circ Syst 11(3):612–626
Luo Y, Chakrabarty K, Ho TY (2012) A cyberphysical synthesis approach for error recovery in digital microfluidic biochips. In: Design, Automation & Test in Europe Conference & Exhibition (DATE), 2012, IEEE, pp 1239–1244
Maftei E, Pop P, Madsen J (2009) Tabu search-based synthesis of dynamically reconfigurable digital microfluidic biochips. In: Proceedings of the 2009 international conference on Compilers, architecture, and synthesis for embedded systems, ACM, pp 195–204
Majumder M, Hansda K, Roy S (2011) A novel single-fault detection technique of digital microfluidic biochip. Int J Comput Sci Appl 1:92–95
Mazutis L, Gilbert JB, Ung WL, Weitz DA, Griffiths AD, Heyman JA (2013) Single-cell analysis and sorting using droplet-based microfluidics. Nat Protocol 8:870–891
Neuži P, Giselbrecht S, Länge K, Jun Huang T, Manz A (2012) Revisiting lab-on-a-chip technology for drug discovery. Nat Rev Drug Discov 11:620–32. https://doi.org/10.1038/nrd3799
Paik P, Pamula VK, Fair RB (2003) Rapid droplet mixers for digital microfluidic systems. Lab Chip 3(4):253–259
Pasqualetti F, Dorfler F, Bullo F (2013) Attack detection and identification in cyber-physical systems. IEEE Trans Autom Contr 58(11):2715–2729
Roy P, Banerjee A (2016) A new approach for root-causing attacks on digital microfluidic devices. In: 2016 IEEE Asian Hardware-Oriented Security and Trust (AsianHOST), IEEE, pp 1–6
Sia SK, Kricka LJ (2008) Microfluidics and point-of-care testing. Lab-Chip 8:1982–1983. https://doi.org/10.1039/B817915H
Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack M, Pamula V (2008) Development of a digital microfluidic platform for point of care testing. Lab Chip 8(12):2091–2104
Su F, Chakrabarty K (2008) High-level synthesis of digital microfluidic biochips. ACM J Emerg Technol Comput Syst (JETC) 3(4):1
Su F, Hwang W, Chakrabarty K (2006) Droplet routing in the synthesis of digital microfluidic biochips. Proceedings of the Design Automation Test in Europe Conference 1:1–6. https://doi.org/10.1109/DATE.2006.244177
Tang J, Ibrahim M, Chakrabarty K, Karri R (2018) Secure randomized checkpointing for digital microfluidic biochips. IEEE Trans Comput-Aid Design Int Circ Syst 37(6):1119–1132
Wang G, Teng D, Fan SK (2011) Digital microfluidic operations on micro-electrode dot array architecture. IET Nanobiotechnol 5:152–160
Wang G, Yi Tse Lai DT, Lu YW, Ho Y, Lee CY (2014) Field-programmable lab-on-a-chip based on microelectrode dot array architecture. IET Nanobiotechnol 8:163–171
Xu T, Chakrabarty K (2007) Integrated droplet routing in the synthesis of microfluidic biochips. In: 2007 44th ACM/IEEE Design Automation Conference, pp 948–953
Xu T, Chakrabarty K (2007) Integrated droplet routing in the synthesis of microfluidic biochips. In: proceedings of the 44th annual Design Automation Conference, pp 948–953
Zhao Y, Xu T, Chakrabarty K (2010) Integrated control-path design and error recovery in the synthesis of digital microfluidic lab-on-chip. ACM J Emerg Technol Comput Syst (JETC) 6(3):11
Zhong Z, Li Z, Chakrabarty K (2017) Adaptive error recovery in meda biochips based on droplet-aliquot operations and predictive analysis. In: 2017 IEEE/ACM International Conference on Computer-Aided Design (ICCAD), pp 615–622, https://doi.org/10.1109/ICCAD.2017.8203834
Zhong Z, Li Z, Chakrabarty K (2018) Adaptive and roll-forward error recovery in meda biochips based on droplet-aliquot operations and predictive analysis. IEEE Trans Multi-Scale Comput Syst 4(4):577–592
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Das, C., Muhuri, S., Chakraborty, S. et al. Security assessment for routing-based synthesis on cyberphysical MEDA-based digital microfluidic biochip. SN Appl. Sci. 3, 721 (2021). https://doi.org/10.1007/s42452-021-04647-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-021-04647-9