Abstract
Mass attenuation coefficients, effective atomic numbers, electron densities and energy absorption and exposure buildup factors for the non-essential and conditionally non-essential amino acids including alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, proline, serine, and tyrosine have been acquired using the latest evaluated photoatomic library of EPICS2017. The library was used by constructing an interpolation script that calculates for all photon attenuation parameters. Comparisons were made using alternative Monte Carlo simulation results for 15 energy points from 59.5 to 1333 keV, and in contrast with experimental works in literature. Good agreements for the mass attenuation coefficients were observed between EPICS2017-based values compared with Monte Carlo code and experimental results. Similar trends for the effective atomic numbers and electron densities were observed from EPICS2017 interpolation and from results found in literature. Conversely, buildup factors acquired by Geometric-Progression fitting parameters were reported in this work preliminarily for most of these biomolecules at different penetration depths. Overall, cysteine showed the most significant deviation among the other non-essential amino acids due to the presence of sulfur in its molecular structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Protein is essential in various biological processes in the human body and plays an important role in the repair, growth, and maintenance of body cells. Proteins can be metabolized and broken down to their building blocks called amino acids (AAs). Each of these AAs can be subcategorized into essential (EAA) or non-essential (NEAA) based on the ability of the body to synthesize the particular AA [1]. In essence, NEAAs are those which can be produced by the human body, while EAAs are those obtained from diet [2]. With the vast roles of the NEAAs including gene expression, cell signaling, neurological processes, acid-base balance, food digestion and absorption, breakdown of nutrients, metabolism, fertilization and immunity, allosteric and epigenetic mechanisms, and tumorigenesis, researches have gained interest in targeting NEAAs for cancer therapy and field of oncology [3, 4].
Radiotherapy in the treatment of malignancy uses high doses of radiation in order to target cancer cells and diminish the size of tumors and slow down the growth and proliferation of cells through DNA damage. To calculate the penetration and dose deposition of X-ray and gamma photons in a particular target, the photon mass attenuation coefficient (\(\mu /\rho\)), effective atomic number, electron density, and buildup factor are important radiological parameters which can be used [5,6,7]. The former quantities are derived from the \(\mu /\rho\) or the cross section. These parameters are significant in the field of radiology and cancer therapy for estimations and optimizations of doses received by the patient.
Accurate values of \(\mu /\rho\) are hence important for biomolecules comprising the human body [8, 9]. The widely used XCOM-NIST [10] can conveniently extract \(\mu /\rho\) for any material for a wide range of photon energies [5, 11,12,13,14]. Alternative methods in literature for calculating \(\mu /\rho\) involve photon transmission experiments and Monte Carlo (MC) simulations [15,16,17,18,19,20,21,22]. A scarcely reported method is by direct interpolations of major libraries other than those available from XCOM-NIST.
This work evaluates for NEAAs the photon attenuation parameters including mass attenuation coefficients, effective atomic numbers, electron densities, and exposure and energy absorption buildup factors, derived using EPICS2017 [23] library for interpolations. The new EPICS2017 library is the official evaluated photon and electron library of the latest ENDF/B-VIII [24]. To compare the mass attenuation coefficients obtained from the EPICS2017, the MC method was used as well as available experimental data found in literature. Exposure and energy absorption buildup factors for the NEAAs were reported in this work at multiple penetration depths using Geometric-Progression (G-P) fitting method.
2 Theory
The mass attenuation coefficient \(\mu /\rho\) (\(\hbox {cm}^{2}\hbox {g}^{-1}\)) for a medium is described by the Beer–Lambert law or Equation (1), where \(I_{0}\) and I are beam incident and transmitted intensities, respectively, \(\rho\) is the density (g \(\hbox {cm}^{-3}\)) of the medium, and x is the thickness or length (cm) [25].
The law of mixtures shown in Equation (2) gives the total mass attenuation coefficients for a multi-element material, which describes it as a linear weighted combination of the \(( \upmu /\uprho ) _{i}\) of its constituent elements, where \(w_{i}\) is weight fraction of the \(i\mathrm{{th}}\) element [26].
The mass attenuation coefficient is subsequently proportional to the total atomic cross section \(\upsigma _{T}\) (\(\hbox {cm}^{2}\) \(\hbox {atom}^{-1}\)) as shown in Equation (3), where \(N_{A}\) is the Avogadro’s number, \(n_{i}\) is the number of atoms in the molecular formula, and \(A_{i}\) is the atomic mass of the \(i\mathrm{{th}}\) element.
The total atomic cross section for each element \(( \upsigma _{T} )_{i}\) is the sum of the partial cross sections for photoelectric, coherent, incoherent, and pair production, described in Equation (4).
The electronic cross section is a quantity described by Equation (5), where \(f_{i}\) is the atom fraction of each element and \(\hbox {Z}_{i}\) is the atomic number of each element [27]. It is related to effective atomic number \(\hbox {Z}_\mathrm{{eff}}\) that is a unitless quantity given by the ratio of total atomic cross section and electronic cross section shown in Equation (6). On the other hand, the electron density \(\hbox {N}_\mathrm{{eff}}\) (electrons \(\hbox {g}^{-1}\)) is related to \(\hbox {Z}_\mathrm{{eff}}\) by Equation (7). Both the \(\hbox {Z}_\mathrm{{eff}}\) and \(\hbox {N}_\mathrm{{eff}}\) are widely used for the calculations of doses in multi-element materials by radiation therapy [28, 29].
The Beer–Lambert law in Equation (1) is only valid for narrow-beam geometry, thin absorbing materials, and monoenergetic photons. In the case of divergences from such conditions, a correction factor called buildup factor B(E, x) accounts for the effects of scattering described in Equation (8). It is a function of both photon energy and penetration depth in terms of the mean free path (mfp) [30]. The mfp is the inverse of the linear attenuation coefficient \(\upmu\) as shown by Equation (9).
Two classifications of the quantity B(E, x) are referred to as the energy absorption buildup factor (EABF) and the exposure buildup factor (EBF), where the quantity of interests are absorbed energy and exposure, respectively [31]. For both classifications of B(E, x), the ANSI/ANS-6.4.3 [32] contains a compilation of G-P fitting parameters (a, b, c, d, \(X_{k}\)) for 23 elements in the photon energy range between 0.015 and 15 MeV, for depth of up to 40 mfp. The use of G-P fitting parameters is an accurate method of estimating buildup factors for different materials at a broad range of energies [33,34,35].
There are three steps for obtaining the EABF and EBF using the ANSI/ANS-6.4.3 G-P fitting parameters: (1) determination of equivalent atomic number \({Z}_\mathrm{{eq}}\) for the material, (2) evaluation of the G-P fitting parameters for the material, and (3) estimation of EABF and EBF using the parameters [30]. The \({Z}_\mathrm{{eq}}\) represents an equivalent atomic number for multi-element materials. It is obtained by considering the ratio of scattering and total attenuation, which monotonically decreases as a function of atomic number [35]. This is described by Equation (10), where R is the ratio of the \(\mu /\rho\) of Compton scattering and the total \(\mu /\rho\), for the material. The \(R_{1}\) and \(R_{2}\) are the ratios of two successive elements with atomic numbers \({Z}_{1}\) and \({Z}_{2}\). The \({Z}_\mathrm{{eq}}\) is used to obtain the G-P fitting parameters of the material. This is done by considering the tabulated G-P fitting parameters of the elements in ANSI/ANS-6.4.3 as illustrated in Equation (11), where F represents each of the five parameters (a, b, c, d, \(X_{k}\)).
Afterward, the term K(E, x) is calculated using the solved G-P fitting parameters through Equation (12). This term represents the photon dose multiplication and change in the shape of the spectrum, with penetration from that at 1 mfp [32]. The buildup factors B(E, x) representing EABF or EBF are then estimated using Equations (13) or (14).
3 Materials and methods
3.1 Non-essential amino acids
All NEAAs investigated include alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, proline, serine, and tyrosine. The chemical constituents of the biomolecules are shown in Table 1. For alanine, arginine, glycine, and proline, experimentally determined mass attenuation coefficients were previously evaluated by Pawar and Bichile [21]. For asparagine, aspartic acid, glutamine, values were recently evaluated by Kore and Pawar [19], and for serine by Gaikwad et al. [16].
3.2 EPICS2017 interpolation
EPICS2017 [23] holds the most recent photoatomic library and is an official library in the latest ENDF/B-VIII release [24]. EPICS2017 is downloadable from the IAEA NDS website in both the ENDL and ENDF data formats. For this work, the version in the ENDF-6 data format was used. The EPICS2017 library was extracted previously for the development of a simple spreadsheet-based program for photon shielding and dosimetry applications [36]. Our previous program was built using MS Excel spline interpolation extensions and additional features via the VBA language. In this present work, we implement the interpolations using an executable Windows-based program.
It is worth mentioning that the predecessors of EPICS2017 are interpolable using the log-log interpolation law. However, due to several users mis-interpolating by using lin-lin, it was decided to convert the new EPICS2017 into a linearized library. Hence, the interpolation in this work was based on the recommended (lin-lin) law [37] and was applied to all partial cross sections, including coherent scattering, incoherent scattering, pair production, triplet production, and photoelectric separately for each subshell. The total atomic cross section was obtained by summing all relevant partial cross sections. Notably, due to the linearization, it is now possible to interpolate the total atomic cross sections also available from the library [38].
3.3 PHITS Monte Carlo method
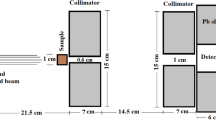
Comparative MC method to determine mass attenuation coefficients was made using PHITS 3.17 [39]. The default EPDL97 (ENDF/B-VII) [40] photoatomic library was used. The modeled geometry is shown in Fig. 1 comprising of a detector covered with a lead shield. This shield was 5-cm thick and had a beam entry diameter of 4 mm for achieving narrow-beam geometry. Each amino acid sample was modeled as a cylinder of 2-cm diameter. Length of each sample was in the range conforming to Nordfors criteria (2 \(\le\) ln(\(I_{0}/I\)) \(\le\) 4) [41]. The surface-to-surface distance between the lead shield and the amino acid sample was fixed at 10 cm.
Photon source was modeled as a monoenergetic pencil beam with 4-mm diameter. The starting energies simulated were 59.5, 81, 88, 122.1, 136.5, 276.4, 302.9, 356, 383.8, 661.7, 834.8, 1173.2, 1252.9, 1274.5 and 1332.5 keV. The [T-Track] tally with energy binning was used to obtain uncollided fluences inside the detector cell. Each run was achieved at a total of \(10^{7}\) simulated source photons per photon energy for each NEAA sample.
4 Results and discussion
An evaluation of the photon attenuation parameters for the NEAAs was made using the EPICS2017 photoatomic library. The results for the mass attenuation coefficients are shown in Tables 2, 3, 4 and 5, which are from EPICS2017 interpolation, from comparative PHITS simulations, and from available experimental values found in literature [16, 19, 21]. It is seen that the PHITS simulation results agree well with EPICS2017 with only marginally lower values. Nonetheless, the difference is owing to the slight dissimilarity in photoatomic libraries, deviation from narrow beam geometry, and relative uncertainties from MC simulation. Importantly, EPICS2017 library has new binding energies translating to new absorption edge energies and cross sections, as compared to the EPDL97 used by the MC method.
On the other hand, experimental results from Pawar and Bichile [21] (alanine, arginine, glycine, and proline) and Gaikwad et al. [16] (serine) showed agreement with the theoretical results. Those obtained by Kore and Pawar [19] for asparagine, aspartic acid, and glutamine were slightly lower than the theoretical results but were nevertheless found to be in similar trends.
Effective atomic numbers and electron densities obtained using EPICS2017 interpolations are shown in Figs. 2 and 3. In the lower energy region in which photoelectric effect is dominant, the values are highest with a negative slope toward the Compton dominated region. For Compton dominated region, both plots tend to dip toward constants and lowest values. The effective atomic numbers in this constant region are between 3.6 and 4.7. These results were also observed for some EAAs reported by Elbashir et al. [15] using the MCNP5 calculations. Notably the MCNP5 is also based on EPDL97. At the constant region, the effective atomic numbers are found to be in descending order of Cys > Asp > Asn > Glu > Ser > Gly > Tyr > Gln > Ala > Pro > Arg. Electron densities at constant region were found to have a narrow spread between 3.2 and 3.3 (\(\times 10^{23}\) electrons \(\hbox {g}^{-1}\)). These values are in descending order of Arg > Ala > Pro > Gln > Ser > Gly > Glu > Asn > Tyr > Cys > Asp. After the constant values at Compton dominated region, rising values are shown due to the increasing probability of pair-production interactions.
EBF and EABF calculated using EPICS2017 and the ANSI/ANS-6.4.3 G-P fitting parameters are shown in Fig. (4) at multiple penetration depths. It is worth noting that buildup factors account for the contribution of scattered photons toward the quantity of interest (i.e., exposure, absorbed energy). The results showed lower EBF and EABF values at the energy regions where scattering is relatively low or insignificant. Since photoelectric effect and pair-productions result in the absorption of the primary photons, these interaction events decrease the likelihood that these photons scatter further within the material. Hence, absorption events generally decrease buildup factors within the medium. EBF and EABF were previously reported for EAAs and some conditional NEAAs [6, 42]. For some EAAs, buildup factors were described with an inverse relation to effective atomic numbers \(\hbox {Z}_\mathrm{{eff}}\) [6]. In this study, the three highest \(\hbox {Z}_\mathrm{{eff}}\) and three lowest showed this inverse relationship with EBF and EABF. This was assessed from the NEAAs at 1 to 40 mfps. It is highlighted that all NEAAs are chemically composed of H, C, N, and O atoms with the exception of cysteine that also contains S atoms. This contributes to cysteine’s larger effective atomic numbers and electron densities at the lower and higher portions of the energy range considered. Consequently, cysteine also demonstrated the lowest values for both types of buildup factors at a large range of photon energies.
5 Conclusion
Photon attenuation parameters of eleven NEAAs have been determined using interpolation method based on the new EPICS2017 library. The EPICS2017 is the current latest library for electron-photon transport and is officially included in ENDF/B-VIII. The EPICS2017 has been interpolated in this work in the recommended lin-lin law using a Windows-based program. Obtained mass attenuation coefficients of NEAAs were compared with MC method results and experimental values in literatures. An agreement with simulation and experimental values was found. Effective atomic numbers and electron densities were also in agreement with those in literature based on MCNP5 calculations. Buildup factors for energy absorption and exposure quantities have been reported via G-P fitting parameters with the use of the EPICS2017 library. Cysteine was found to have a significantly larger effective atomic number which translates to significantly lower buildup factor values, owing to the S atom comprising its chemical structure. These results can serve in the fields of radiology and medical dosimetry. Furthermore, the interpolation method using EPICS2017 was found to be a useful and recent alternative to the generally used XCOM-NIST.
References
Reeds PJ (2000) Dispensable and indispensable amino acids for humans. J Nutr 130(7):1835S-1840S. https://doi.org/10.1093/jn/130.7.1835S
Wu G (2010) Functional amino acids in growth, reproduction, and health. Adv Nutr 1(4):31–37. https://doi.org/10.3945/an.110.1008.1
Hou Y, Yin Y, Wu G (2015) Dietary essentiality of “nutritionally non-essential amino acids” for animals and humans. Experimental Biology and Medicine 240(8):997–1007. https://doi.org/10.1177/1535370215587913
Choi BH, Coloff JL (2019) The diverse functions of non-essential amino acids in cancer. Cancers. https://doi.org/10.3390/cancers11050675
Bursalıoğlu EO, İçelli O, Balkan B, Kavanoz HB, Okutan M (2014) Photon atomic parameters of nonessential amino acids for radiotherapy and diagnostics. J Amino Acids. https://doi.org/10.1155/2014/434519
Bursalıoğlu E, Balkan B, Kavanoz HB, Okutan M, Içelli O, Yalçin Z (2014) Energy absorption and exposure buildup factors of essential amino acids. BioMed Res Int. https://doi.org/10.1155/2014/359754
Kurudirek M, Medhat ME (2014) Robust determination of mass attenuation coefficients of materials with unknown thickness and density. Radiat Phys Chem 100:65–69. https://doi.org/10.1016/j.radphyschem.2014.03.031
Tonguc BT, Arslan H, Al-Buriahi MS (2018) Studies on mass attenuation coefficients, effective atomic numbers and electron densities for some biomolecules. Radiat Phys Chem 153:86–91. https://doi.org/10.1016/j.radphyschem.2018.08.025
Al-Buriahi MS, Arslan H, Tonguc BT (2019) Investigation of photon energy absorption properties for some biomolecules. Nuclear Sci Tech. https://doi.org/10.1007/s41365-019-0636-9
Berger MJ, Hubbell JH, Seltzer SM, Chang J, Coursey JS, Sukumar R, Zucker DS, Olsen K (2010) XCOM: Photon cross section database (version 1.5). National Institute of Standards and Technology, Gaithersburg, MD. https://doi.org/10.18434/T48G6X
Ahmadi M, Lunscher N, Yeow JTW (2013) Effective atomic numbers and electron densities of bacteriorhodopsin and its comprising amino acids in the energy range 1 keV–100 GeV. Nuclear Inst Methods Phys Res B 300:30–34. https://doi.org/10.1016/j.nimb.2013.01.002
Manohara SR, Hanagodimath SM (2007) Studies on effective atomic numbers and electron densities of essential amino acids in the energy range 1 keV–100 GeV. Nuclear Instrum Methods Phys Res Sect B Beam Interact Mater Atoms 258(2):321–328. https://doi.org/10.1016/j.nimb.2007.02.101
Manohara SR, Hanagodimath SM (2007) Effective atomic numbers for photon energy absorption of essential amino acids in the energy range 1 keV–20 MeV. Nuclear Instrum Methods Phys Res Sect B Beam Interact Mater Atoms 264(1):9–14. https://doi.org/10.1016/j.nimb.2007.08.018
Manohara SR, Hanagodimath SM, Gerward L (2009) The effective atomic numbers of some biomolecules calculated by two methods: a comparative study. Med Phys 36(1):137–141. https://doi.org/10.1118/1.3030952
Elbashir BO, Dong MG, Sayyed MI, Issa SA, Matori KA, Zaid MH (2018) Comparison of Monte Carlo simulation of gamma ray attenuation coefficients of amino acids with XCOM program and experimental data. Results Phys 9:6–11. https://doi.org/10.1016/j.rinp.2018.01.075
Gaikwad DK, Pawar PP, Selvam TP (2017) Mass attenuation coefficients and effective atomic numbers of biological compounds for gamma ray interactions. Radiat Phys Chem 138:75–80. https://doi.org/10.1016/j.radphyschem.2017.03.040
Gowda S, Krishnaveni S, Gowda R (2005) Studies on effective atomic numbers and electron densities in amino acids and sugars in the energy range 30–1333 keV. Nuclear Instrum Methods Phys Res Sect B Beam Interact Mater Atoms 239(4):361–369. https://doi.org/10.1016/j.nimb.2005.05.048
Hila FC, Dicen GP, Javier-Hila AMV, Asuncion-Astronomo A, Guillermo NRD, Rallos RV, Navarrete IA, Amorsolo AV (2020) Determination of photon shielding parameters for soils in mangrove forests, Philippine J Sci(In-Press)
Kore PS, Pawar PP (2014) Measurements of mass attenuation coefficient, effective atomic number and electron density of some amino acids. Radiat Phys Chem 98:86–91. https://doi.org/10.1016/j.radphyschem.2013.12.038
More CV, Lokhande RM, Pawar PP (2016) Effective atomic number and electron density of amino acids within the energy range of 0.122–1.330 MeV. Radiat Phys Chem 125:14–20. https://doi.org/10.1016/j.radphyschem.2016.02.024
Pawar PP, Bichile GK (2013) Studies on mass attenuation coefficient, effective atomic number and electron density of some amino acids in the energy range 0.122–1.330 MeV. Radiat Phys Chem 92:22–27. https://doi.org/10.1016/j.radphyschem.2013.07.004
Sahin A, Bozkurt A (2019) Monte Carlo calculation of mass attenuation coefficients of some biological compounds. Süleyman Demirel Univ Faculty Arts Sci J Sci 14(2):408–417
Cullen DE (2018) A survey of photon cross section data for use in EPICS2017, IAEA-NDS-225, rev. 1
Brown DA, Chadwick MB, Capote R, Kahler AC, Trkov A, Herman MW, Sonzogni AA, Danon Y, Carlson AD, Dunn M, Smith DL, Hale GM, Arbanas G, Arcilla R, Bates CR, Beck B, Becker B, Brown F, Casperson RJ, Conlin J, Cullen DE, Descalle MA, Firestone R, Gaines T, Guber KH, Hawari AI, Holmes J, Johnson TD, Kawano T, Kiedrowski BC, Koning AJ, Kopecky S, Leal L, Lestone JP, Lubitz C, Márquez Damián JI, Mattoon CM, McCutchan EA, Mughabghab S, Navratil P, Neudecker D, Nobre GP, Noguere G, Paris M, Pigni MT, Plompen AJ, Pritychenko B, Pronyaev VG, Roubtsov D, Rochman D, Romano P, Schillebeeckx P, Simakov S, Sin M, Sirakov I, Sleaford B, Sobes V, Soukhovitskii ES, Stetcu I, Talou P, Thompson I, van der Marck S, Welser-Sherrill L, Wiarda D, White M, Wormald JL, Wright RQ, Zerkle M, Žerovnik G, Zhu Y (2018) ENDF/B-VIII.0: The 8th major release of the nuclear reaction data library with CIELO-project cross sections, new standards and thermal scattering data. Nuclear Data Sheets 148:1–142. https://doi.org/10.1016/j.nds.2018.02.001
Al-Buriahi MS, Mann KS (2019) Radiation shielding investigations for selected tellurite-based glasses belonging to the TNW system. Mater Res Express 6(10):105206. https://doi.org/10.1088/2053-1591/ab3f85
Matthews F, Rawlings R (1999) Non-destructive testing, In: Composite Materials, Elsevier, pp 415–447. https://doi.org/10.1016/b978-1-85573-473-9.50018-7
Al-Buriahi MS, Tonguc BT (2020) Mass attenuation coefficients, effective atomic numbers and electron densities of some contrast agents for computed tomography. Radiat Phys Chem. https://doi.org/10.1016/j.radphyschem.2019.108507
Akman F, Durak R, Turhan MF, Kaçal MR (2015) Studies on effective atomic numbers, electron densities from mass attenuation coefficients near the K edge in some samarium compounds. Appl Radiat Isot 101:107–113. https://doi.org/10.1016/j.apradiso.2015.04.001
Singh T, Kaur P, Singh PS (2007) A study of photon interaction parameters in some commonly used solvents. J Radiol Protect 27(1):79–85. https://doi.org/10.1088/0952-4746/27/1/005
Olarinoye IO, Odiaga RI, Paul S (2019) EXABCal: A program for calculating photon exposure and energy absorption buildup factors. Heliyon. https://doi.org/10.1016/j.heliyon.2019.e02017
Sayyed MI, AlZaatreh MY, Dong MG, Zaid MH, Matori KA, Tekin HO (2017) A comprehensive study of the energy absorption and exposure buildup factors of different bricks for gamma-rays shielding. Results Phys 7:2528–2533. https://doi.org/10.1016/j.rinp.2017.07.028
ANSI/ANS 6.4.3-1991 (1991) Gamma-ray attenuation coefficients and buildup factors for engineering materials, ANSI/ANS 6.4.3-1991, ANSI/ANS-6.4.3. American Nuclear Society, La Grange Park, Illinois
Harima Y (1983) An approximation of gamma-ray buildup factors by modified geometrical progression. Nuclear Sci Eng 83(2):299–309
Harima Y, Sakamoto Y, Tanaka S, Kawai M (1986) Validity of the geometric-progression formula in approximating gamma-ray buildup factors. Nuclear Sci Eng 94(1):24–35
Harima Y (1993) An historical review and current status of buildup factor calculations and applications. Radiat Phys Chem 41(4–5):631–672. https://doi.org/10.1016/0969-806x(93)90317-n
Hila FC, Amorsolo AV, Javier-Hila AMV, Guillermo NRD (2020) A simple spreadsheet program for calculating mass attenuation coefficients and shielding parameters based on EPICS2017 and EPDL97 photoatomic libraries, Radiat Phys Chem 109122 https://doi.org/10.1016/j.radphyschem.2020.109122
Cullen DE (2019) EPICS2017: April 2019 status report, IAEA-NDS-228
Cullen DE (2010) Nuclear data preparation. In: Handbook of nuclear Engineering, Springer US, pp. 279–425. https://doi.org/10.1007/978-0-387-98149-9_4
Sato T, Iwamoto Y, Hashimoto S, Ogawa T, Furuta T, Ichiro Abe S, Kai T, Tsai PE, Matsuda N, Iwase H, Shigyo N, Sihver L, Niita K (2018) Features of particle and heavy ion transport code system (PHITS) version 3.02. J Nuclear Sci Technol 3131:1–7. https://doi.org/10.1080/00223131.2017.1419890
Cullen D, Hubbell J, Kissel L (1997) EPDL97: the evaluated photon data library, UCRL-50400 and Vol. 6 and Rev. 5 6, 1–35
Creagh DC, Hubbell JH (1990) Problems associated with the measurement of X-ray attenuation coefficients. II. Carbon. Report on the international union of crystallography X-ray attenuation project, Acta Crystallographica Sect A Found Crystallogr 46(5):402–408. https://doi.org/10.1107/s0108767389013887
Kurudirek M, Özdemir Y (2011) A comprehensive study on energy absorption and exposure buildup factors for some essential amino acids, fatty acids and carbohydrates in the energy range 0.015-15 MeV up to 40 mean free path, Nuclear Inst. Methods Phys Res B 269(1):7–19. https://doi.org/10.1016/j.nimb.2010.10.015
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Javier-Hila, A.M.V., Javier, B.C.V., Hila, F.C. et al. Photon attenuation parameters of non-essential amino acids using EPICS2017 library interpolations. SN Appl. Sci. 3, 542 (2021). https://doi.org/10.1007/s42452-021-04507-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-021-04507-6