Abstract
Excitation-emission matrix fluorescence (EEMF) spectroscopy was combined with orthogonal signal correction (OSC) assisted partial least square (PLS) analysis to achieve simultaneous quantification of aqueous mixtures of carcinogenic and mutagenic polycyclic aromatic hydrocarbons (PAHs). The application of OSC algorithm on EEMF data sets prior to PLS analysis was found to significantly improve the performance of calibration model towards the PAHs quantification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The careless handling of the petroleum products are mainly responsible for the presence of polycyclic aromatic hydrocarbons (PAHs) in water samples [1,2,3,4,5]. PAHs are carcinogenic and mutagenic in nature and their regular monitoring in water samples is an important task to avoid serious health concern [1,2,3,4,5]. Gas or liquid phase chromatography coupled with either mass spectrometry or UV–Visible spectrophotometers are the commonly used techniques for the quantification of PAHs in water samples [6,7,8,9]. The chromatographic analysis are costly and involve laborious sample preparation steps. As a result, the chromatographic techniques less preferred especially in the developing countries where PAHs monitoring needs to be carried out on routine basis.
PAHs have the rigid molecular framework making them highly fluorescent in nature [5, 10,11,12,13,14,15]. Thus, fluorescence a simple and sensitive technique could be a method of choice for the routine analysis of PAHs in water samples. Excitation-emission matrix fluorescence (EEMF) is multi-parametric fluorescence techniques that simultaneously capture the fluorescence activity of all the fluorescent molecules in a single plot [10, 16,17,18,19]. EEMF spectrum of a fluorescent molecule depicts the variation of fluorescence intensity as the excitation (λex) and emission (λem) wavelengths are varied. The modern fluorimeters allows the acquisition of EEMF data in a fast and automatic manner over the user specified parameters. The modern fluorimeters are also equipped with appropriate software that enables auto-removal of Rayleigh scattering signals (1st and 2nd order) at the data acquisition level. The fluorescence intensity for a mixture of fluorescent components in EEMF spectroscopy corresponds to the summation of individual fluorescence intensities of each components weighted by their corresponding concentrations [10, 16,17,18,19]. EEMF data sets can be analyzed to develop a calibration model provided one could correlate the intensity of the fluorescent components of the mixture with their corresponding concentrations.
Partial least square (PLS) analysis [20,21,22,23] is one of the most commonly used technique to develop a calibration model. PLS algorithm essentially finds a set of factors (also known as latent variables) that not only describe the maximum variation of the spectral data set but also maximizes the correlation between the spectral and concentration data matrix [20,21,22,23]. However, PLS model of highly correlated spectral data sets generated by the modern fluorimeters are harder to interpret. PLS model for such data sets require more number of factors for modeling. The PLS model of such data captures the maximum spectral variation but despite using more number of factors often fails to correlate the spectral and concentration related information. Often, PLS modeling of spectral data sets goes against the principle of parsimony [20] that states the model must be least complex. In order to achieve the maximum correlation between the spectral and concentration data matrices, it is suggested that one could apply orthogonal signal correction (OSC) technique [24,25,26,27] prior to PLS analysis. The OSC technique essentially uses principal component analysis (PCA) technique [20, 27] to removes a set of factors explaining the maximum variations of the spectral data matrix that are orthogonal to the concentration matrix. The simplified spectral data set along with concentration data matrix can further be processed by PLS algorithm to develop a robust and parsimonious calibration model.
In literature, EEMF combined with second-order calibration methods such as parallel factor (PARAFAC) [12, 14, 16] and alternate trilinear decomposition (ATLD) [14, 15] methods have been successfully used for the analysis of PAHs mixtures even in the presence of unknown interferences. These methods involve decomposition of the spectral data sets without involving any concentration related information. As a result, often the correlation between the spectral intensity and the concentration is not achieved to the desired level. Whereas, the objective of the PLS algorithm is to maximize this correlation so that a robust model could be obtained. In addition to this, application of OSC prior to PLS further improve the correlation by removing the information from the spectral data sets that are not correlated with concentration data matrix. Thus, the application of OSC assisted PLS analysis could be an attractive proposition for developing robust calibration model for the quantification of PAHs having significant spectral overlap with each other.
The objective of the present work is to take the advantages associated with the OSC assisted PLS algorithm for analyzing large volume of EEMF spectroscopic data sets and propose a simple and sensitive analytical procedure for achieving the simultaneous quantification of aqueous mixture of five PAHs without pre-separation. To carry out the present work, aqueous mixtures anthracene, benzo[a]pyrene, chrysene, pyrene and fluoranthene were taken as the test case. It was a thoughtful choice to select the overlapping PAHs that makes the analysis. Each of the selected PAHs have diverse molecular structure, for example, anthracene is composed of three fused benzene ring, both chrysene and pyrene are composed of four fused benzene ring with different arrangement, benzo[a]pyrene is composed of five fused benzene ring whereas fluoranthene is composed of naphthalene and benzene unit connected by five membered ring [1,2,3,4,5]. Moreover, chrysene and benz[a]pyrene are known to be the most carcinogenic and mutagenic PAHs [1,2,3,4,5]. Each of the selected PAHs are known to penetrate the skin and they are highly soluble in lipids causing serious damage to vital organs of both human and animals [1,2,3,4,5]. In addition to this, the selected PAHs have the significant spectral overlap among each other that could really test the OSC assisted PLS algorithm. To the best of our knowledge, it is first report that combines OSC assisted PLS algorithm with EEMF technique towards the simultaneous quantification of aqueous mixture of five potential carcinogenic and mutagenic PAHs having significant spectral overlap with each other. It is believed that proposed approach would be useful towards making simple and fast analysis of these PAHs in water samples on the routine basis.
2 Material and methods
2.1 Chemicals and sample preparation
Each of the five PAHs, anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene were obtained from the Aldrich. Separate stock solutions were prepared by dissolving 20 mg of anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene in 200 ml of methanol. A calibration set of 20 samples and a validation set of 5 samples were prepared by pipetting random volumes of these PAHs from their respective stock solutions. The methanol was removed by passing the inert gas followed by dilution with distilled water. The final concentrations of PAHs in aqueous mixtures of calibration and validation set samples are summarized in Table 1.
2.2 Data acquisition
EEMF data sets for aqueous mixtures of five PAHs anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene were acquired by the following parameters.
-
(i)
Excitation wavelength range: 270–450 nm with a step size of 5 nm
-
(ii)
Emission wavelength range: 300–570 nm with a step size of 2 nm
-
(iii)
Excitation slit width: 5 nm
-
(iv)
Emission slit width: 5 nm
2.3 Computational platform
The data arrangement, data plotting and analysis were carried out using MATLAB software.
2.4 Orthogonal signal correction algorithm
The OSC algorithm [24,25,26,27] removes the major variations from the spectral data matrix X that are not correlated with the concentration data matrix Y. Application of OSC as a pre-processing technique prior to PLS help in maximizing the correlation between the spectral and concentration data matrices and hence improves the quantification accuracy of the developed calibration model. The OSC algorithm could be explained using following steps:
-
(i)
X = TPT
-
(ii)
tn = t1 − Y(YTY)-1Yt1
-
(iii)
W = X−1tn
-
(iv)
t = XW
-
(v)
PT = tTX/(ttn)
-
(vi)
Xcorr = X − tPT.
The Xcorr can be used as the spectral data matrix input for the PLS algorithm [20,21,22,23].
3 Results and discussion
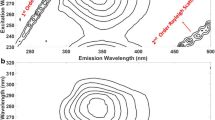
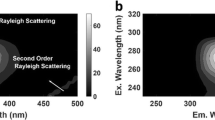
Pure EEMF spectral profiles of anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene are shown as contour map in Fig. 1. It can be seen that selected PAHs have the significantly overlapped EEMF spectral profiles. Thus, it can be seen that designed calibration set is a challenging system to analyze and can test the proposed analytical approach. To show the novelty of the proposed approach, the present work is divided in two segments. In the first step, traditional PLS algorithm is applied. In the second step, OSC assisted PLS algorithm is used to analyze EEMF data sets and the obtained results of these two steps are compared using appropriate statistical parameters.
3.1 Data arrangements and finding the optimum number of factors
EEMF data sets were arranged as two-way arrays of dimensions sample × (λem × λex) for both calibration and validation sets. The dimension of calibration data set (X) is 20 × 4760 (= 136 × 35) and dimension of validation set is 5 × 4760 (= 136 × 35). The concentration related information of calibration set was arranged in a matrix (Y) of dimension 20 × 5 (sample × analytes). The first, second, third, fourth and fifth columns of Y contains the concentration of anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene, respectively. Before proceeding with data analysis, both calibration and validation data sets were blank subtracted to mitigate the influence of Raman scattering. As there are five PAHs in the mixture, thus going by the principle parsimony one must use five factors one for each chemical component to carry out the PLS analysis. Thus, in order to ensure that there is true correlation between the chemical and mathematical ranks of the EEMF data sets, PCA associated with leave one out cross validation (LOOCV) [20, 27] was used. The root mean square errors of cross-validation (RMSECV) [20, 27] value against the number of factor plot, shown in Fig. 2, clearly show that PCA model of five factor achieves the minimum error, thus, the choice of five factor one for each molecule is justified.
3.2 PLS analysis on EEMF data sets of aqueous mixtures of PAHs
The suitably arranged EEMF data set X of dimension 20 × 4760 of calibration set along with concentration data matrix Y of dimension 20 × 5 was subjected to PLS analysis with five factors. The data sets are mean-centered across the samples prior to analysis. The developed model explains the 95% variance of X block and 58% variance of Y block. The regression equations relating the actual and calibrated concentration for each of the five PAHs are summarized in Table 2. The square of the correlation coefficient (R2) [20, 27], Root mean square of error of calibration (RMSEC) [20, 27] values for the PLS model are reported in Table 2. The developed PLS model was further used to analyze the samples of validation set. The calculated root mean square error of prediction (RMSEP) [20, 27] value for the validation set are reported in Table 2. The slope of the regression equations were found to vary in the range 0.60–0.72 suggesting the PLS model is under estimating each of the five PAHs; ideally, the slope of these regression equations must be close to unity. The R2 values were also found to vary in the range of 0.6–0.7 suggesting the actual and predicted concentrations are not correlated with each other to the desired level sought while quantifying such harmful chemicals. The inadequacy of the PLS model is also reflected in the relatively high RMSEC values varying in the range 30–40% and RMSEP values varying in the range 35–50% for the analyzed PAHs. The evaluation of all these parameters clearly suggest that developed PLS model is far off and it could mainly be because the model failed to capture the Y block (concentration) relation variations. It can further be attributed to the fact that the selected PAHs have the significant spectral overlap with each other that probably obstruct the maximization of the correlation between the concentration and spectral data matrix. To see if it is possible to improve the model, OSC assisted PLS modeling is carried in the next step and the obtained results are compared.
3.3 OSC assisted PLS analysis on EEMF data sets of aqueous mixtures of PAHs
The one possibility to increase the correlation between the spectral and concentration data matrices is to remove the factors from the PLS models that are orthogonal to concentration information. It can be achieved using the OSC algorithm as preprocessing technique prior to PLS analysis. The EEMF data sets X of dimensions 20 × 4760 of calibration set was subjected to OSC algorithm for simplifying the data set. The algorithm finds the first principal component of X data set using PCA. In the next step, the loading vector is rotated so that the scores are orthogonal to concentration data matrix Y of dimension 20 × 5. The rotated loading vector approximates the spectral features that are not influenced by the changes in the property of interest described in the concentration matrix. The OSC processed spectral data sets along with concentration data matrix Y is subjected to PLS analysis with five factors. The developed model was found to explain the 99.5% variance of X block and 100% variance of Y block. It can be seen that there was a significant improvement in the variance captured by the PLS model for the concentration data set. The regression equations relating the actual and calibrated concentration of each of the five PAHs are summarized in Table 3. The slope of the regression equations were found to vary in the range 0.9–0.95 clearly showing a significant improvement in the performance of calibration model. The R2 for each of the analyzed PAHs are summarized in Table 3. The R2 values, reported in Table 3, were also found to vary in the range of 0.9–0.95 suggesting very good correlation between the actual and predicted concentrations for each of the five PAHs. A significant improvement could also be seen in the RMSEC values for each of the five PAHs, the RMSEC values were found to vary in the range of 3–8%. The developed OSC assisted PLS model was further tested by using an external validation set of five samples. The weights and the loading vectors obtained from OSC step of calibration set were used to remove the information from the EEMF data sets of the validation set that are orthogonal to concentration data set. The OSC corrected EEMF data sets are further subjected to the PLS calibration model. The RMSEP values for each of the five PAHs, given in Table 3, were found to vary in the rage of 5–10%. A significant improvement in the PLS model performance could be seen even while analyzing the samples of external validation sets.
Overall obtained results clearly suggest that application of OSC algorithm prior to PLS analysis significantly improves the performance of calibration model for each of the five PAHs even if they have the significant spectral overlap with each other. The application of OSC algorithm helped in removing the information from the spectral data sets that are not correlated with the concentration related information. The OSC assisted PLS analysis of EEMF spectroscopic data sets provide a viable analytical approach for the quantification of aqueous mixtures of carcinogenic and mutagenic PAHs.
4 Conclusions
The present work reports that application of OSC algorithm on EEMF data sets prior to PLS analysis can significantly improve the performance of calibration model by removing the information form the spectral data that are nor correlated with concentration data matrix. The usefulness of the proposed approach is successfully demonstrated by analyzing the aqueous mixtures of five PAHs including anthracene, benzo[a]pyrene, chrysene, fluoranthene and pyrene having significant spectral overlap with each other. The obtained results of the present work clearly showed that OSC assisted PLS along with EEMF provides a simple and sensitive analytical means of quantifying the carcinogenic and mutagenic PAHs in water samples.
References
Biorseth A (1983) Handbook of polycyclic aromatic hydrocarbons. Marcel Decker, New York, p 1983
Spier CR, Vadas GG, Kaattari SL, Unger MA (2011) Near real-time, on-site, quantitative analysis of PAHs in the aqueous environment using an antibody-based biosensor. Environ Toxicol Chem 30:1557–1563
Vo-Dinh T (1989) Chemical analysis of polycyclic aromatic compounds. Wiley-Interscience, New York
Neff JM (1979) Polycyclic aromatic hydrocarbons in the aquatic environment: sources, fate and biological effects. Applied Sciences, London
Kumar K, Mishra AK (2011) Simultaneous quantification of dilute aqueous solutions of certain polycyclic aromatic hydrocarbons (PAHs) with significant fluorescent spectral overlap using total synchronous fluorescence spectroscopy (TSFS) and N-PLS, unfolded-PLS and MCR-ALS analysis. Anal Methods 3:2616–2624
Manoli E, Samara C (1999) Polycyclic aromatic hydrocarbons in natural waters: sources, occurrence and analysis. Trends Anal Chem 18:417–428
Kiss GZ, Puchony V, Hlavay J (1996) Determination of polycyclic aromatic hydrocarbons in precipitation using solid phase extraction and column liquid chromatography. J Chromatogr A 725:261–272
Bourdat-Deschamps M, Daudin JJ, Barriuso E (2007) An experimental design approach to optimise the determination of polycyclic aromatic hydrocarbons from rainfall water using stir bar sorptive extraction and high performance liquid chromatography-fluorescence detection. J Chromatogr A 1167:43–53
Portet-Koltalo F, Oukebdane K, Robin L, Dionnet F, Desbene PL (2007) Quantification of volatile PAHs present at trace levels in air flow by aqueous trapping-SPE and HPLC analysis with fluorimetric detection. Talanta 71:1825–1833
Kumar K, Tarai M, Mishra AK (2017) Unconventional steady-state fluorescence spectroscopy as an analytical technique for analyses of complex-multifluorophoric mixtures. Trends Anal Chem 97:216–243
Wang H, Zhang Y, Xiao X (2010) Quantification of polycyclic aromatic hydrocarbons in water: a comparative study based on three-dimensional excitation-emission matrix fluorescence. Anal Sci 26:1271–1276
Gu HW, Zhang SH, Wu BC, Chen W, Wang JB, Liu Y (2018) A green chemometrics-assisted fluorimetric detection method for the direct and simultaneous determination of six polycyclic aromatic hydrocarbons in oil-field wastewaters. Spectrochim Acta A 200:93–101
Yang RJ, Dong GM, Sun XS, Yang YR, Yu YP, Liu HX, Zhang WY (2018) Feasibility of the simultaneous determination of polycyclic aromatic hydrocarbons based on two dimensional fluorescence correlation spectroscopy. Spectrochim Acta A 190:342–346
Yang Z, Liu T, Wang Y, Yuan Y, Shang F (2018) Comparison of several third-order correction algorithms applied to fluorescence excitation–emission-sample data array: Interference-free determination of polycyclic aromatic hydrocarbons in water pollution. Spectrochim Acta A 205:381–390
Wang HB, Zhang YJ, Xiao X, Yu SH, Liu WQ (2011) Application of excitation–emission matrix fluorescence combined with second-order calibration algorithm for the determination of five polycyclic aromatic hydrocarbons simultaneously in drinking water. Anal Methods 3:688–695
Rho JH, Stuart JL (1978) Automated three-dimensional plotter for fluorescence measurements. Anal Chem 50:620–625
Freegarde M, Hatchard CG, Parker CA (1971) Oil spilt at sea: its identification, determination, and ultimate fate. Lab Pract 20:35–40
Warner IM, Callis JB, Davidson ER, GoutermannM CGD (1975) Fluorescence analysis: a new approach. Anal Lett 8:665–681
Kumar K, Mishra AK (2013) Analysis of dilute aqueous multifluorophoric mixtures using excitation-emission matrix fluorescence (EEMF) and total synchronous fluorescence (TSF) spectroscopy: a comparative evaluation. Talanta 117:209–220
Kramer R (1998) Chemometric techniques for quantitative analysis. Marcel Dekker, New York
Wold S, Ruhe A, Wold H, Dunn WJ (1984) The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J Sci Stat Comput 5:735–743
Geladi P, Kowalski B (1986) Partial least square regression: a tutorial. Anal Chim Acta 185:1–17
Lorber A, Wangen LE, Kowalski BR (1987) A theoretical foundation for the PLS algorithm. J Chemometr 1:19–31
Sjoblom J, Svensson O, Josefson M, Kullberg H, Wold S (1998) An evaluation of orthogonal signal correction applied to calibration transfer of near infrared spectra. Chemometr Intell Lab Syst 44:229–244
Kim K, Lee JM, Lee I (2005) A novel multivariate regression approach based on kernel partial least squares with orthogonal signal correction. Chemometr Intell Lab Syst 79:22–30
Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemometr 16:119–128
Wise BM, Gallaghar NB, Bro R, Shaver JM (2006) PLS_Toolbox 4.0, Eigenvector research, USA
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author Keshav kumar declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, K. Orthogonal signal correction assisted PLS analysis of EEMF spectroscopic data sets: fluorimetric analysis of polycyclic aromatic hydrocarbon mixtures. SN Appl. Sci. 2, 831 (2020). https://doi.org/10.1007/s42452-020-2665-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2665-8