Abstract
The two adiponectin receptors (AdipoR1 and AdipoR2) have been implicated in glucose and lipid regulation involved in several metabolic pathologies including type II diabetes. Their exact biochemical functions and mechanisms remain poorly understood. Moreover, these receptors do not yet have data on possible co-crystallized active ligands. In this study, we applied different computational methodologies to address three main unanswered questions: first, the localization and validation of possible binding sites; second, the generation of novel ligands with amenable characteristics to target the receptors; and third, the determination of important chemical interactions between the ligands and the receptors. Computational analysis of the binding site reveals that the residues triad R267, F271, and Y310 could be responsible for changes in the spatial arrangement and geometry of the binding pocket in AdipoR1. Molecular docking results in high docking scores of − 13.6 and − 16.5 kcal/mol for the top best ligands in AdipoR1 and AdipoR2 respectively. Finally, molecular dynamics suggests that hydrolytic activity may be possible with these compounds and that this reaction could be mediated by aspartic acid residues. The two adiponectin receptors have an endogenous protein ligand, adiponectin. However the synthesis is expensive and technically challenging. Although some debatable agonists have been proposed investigations of suitable synthetic ligands are indeed, very much needed for targeting these receptors and their associate pathologies and metabolic pathways. Furthermore, these findings provide a framework for further biochemical investigations of amenable compounds for drug discovery in order to target these receptors and their associated pathologies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Adiponectin is an adipokine involved in several pathologies associated with the regulation of glucose and the oxidation of fatty acids [1,2,3,4,5]. A diminished plasma level of the protein has been associated with increased obesity in human studies [6, 7]. Several reports have shown the beneficial pleiotropic activity of this protein in pathologies including type II diabetes, dyslipidemia, and the metabolic syndrome but also in conditions of the immune system (common variable immune deficiency, CVID) and cancer [1, 5, 8,9,10,11,12]. Animal studies have shown that the augmentation of adiponectin correlates with improved insulin sensitivity and glucose management in mice [13, 14]. The adiponectin receptors (AdipoRs: AdipoR1 and AdipoR2) are key components in mediating the activity of adiponectin [4, 14]. The two receptors belong to the progesterone adipoQ receptor (PAQR) family in the superfamily of rhodopsin-like receptors and pumps. This family includes seven transmembrane (7TM) G-protein-coupled receptors (GPCRs) [15]. AdipoRs, like GPCRs, have 7TM architecture however, they present a unique conformation with associated distinct properties from GPCRs and other close families [3, 4, 8, 15, 16]. Specifically, AdipoRs present an extracellular C-terminus and a cytosolic N-terminus, overall exhibiting a topology that is opposite to all known GPCRs [4]. These two receptors induce the downstream activation of distinct pathways, namely the AMPK and the PPAR-α for AdipoR1 and AdipoR2 respectively [17]. However, this is, debatably, not exhaustive for the biochemical routes that these receptors activate [18,19,20,21]. Further evidence has highlighted their probable independent ceramidase activity and, at the present time, more evidence has emerged regarding this activity in AdipoRs [9, 14, 22].
The crystal structures of the two receptors reveal a common zinc metal ion adjacent to the intracellular portion of the proteins and an extensive internal cavity spanning almost the full length of the proteins, from the extracellular lipid bilayer to the internal cytosolic space as depicted in Fig. 1 [3, 14]. A probable catalytic site for ceramidase activity has been recently characterized as a part of the central cavity with the zinc ion implicated in modulating free fatty acid (FFA) interactions [14, 23]. Electron density maps of the central cavity indicate that this may be a key modulating feature for molecules’ accessibility to the two receptors [14].
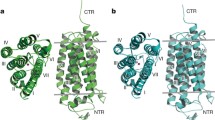
Adiponectin receptors PDBs: 5LXG and 5LX9. a Overlay of the AdipoR1 and 2 showing the difference in overall spatial conformation. b Detailed view of the arrangements of the TM5 and ICL2 secondary structures. c Extracellular view of the transmembrane secondary structures of 5LXG and 5LX9 receptors. d Representation of the central cavity (in grey) generated by KVFinder [24]
A third receptor, T-cadherin, exists with specific affinity for adiponectin in smooth muscle and endothelial cells but lacking in hepatocytes where adiponectin is most active [25, 26]. However, the exact pathophysiological relevance between adiponectin and T-cadherin in several pathologies including diabetes remains unclear [25,26,27,28]. Due to the different specific structure of T-cadherin and its pathological relevance the analysis of this protein is out of the scope of this paper and may be considered in our future work.
At present, neither the co-crystallization nor binding sites localization and/or characterization has been described. Moreover only one synthetic agonist (AdipoRon) has been described however, it has shown high dissociation constant (Kd), thus a low binding affinity, in the range of 1.8 and 3.1 μM (Kd) for adipoR1 and AdipoR2 respectively [29]. This presents an opportunity for the examination of novel molecules and computational analysis of the AdipoRs.
In the context of computational analysis, the conformational changes of at least AdipoR1 (two crystallographic structures in open and closed conformations) suggests that metastable conformations may occur and, therefore, a comprehensive approach that includes an analysis of the conformational changes may be important for ligand recognition, site identification, and validation [14, 30]. The initial clues for possible binding pockets have been described in the literature due to the presence of oleic acid in the central cavity of AdipoR2 (PDB: 5LX9), which has been used to describe the ceramidase activity of these receptors. From this initial evidence, we have addressed three main issues regarding these receptors: binding pocket location, binding pocket validation, and ligand-receptor dynamics. The prediction of possible binding regions was explored by using SiteMap (Schrödinger, LLC, NY, USA) [31,32,33] and intrinsic dynamic domain (IDD)-based analysis [34, 35]. This methodology employs an elastic network model (ENM) that considers its Gaussian and anisotropic network [36, 37]. Next, we generated suitable ligands for these receptors. The initial library of ligands was chosen with the notion that the structural chemical space is immensely large and that biologically active compounds constitute separate closed groups; thus, the analysis of diverse compounds is more prone to produce less useful leads than closely-related compounds [38, 39]. From an initial set of scaffolds, more compounds were retrieved in PubChem using manual and automated searches. Finally, we analyzed the molecular simulations to determine possible common mechanisms of action between the ligands. Some mechanisms of action, conserved among different classes of proteins and hydrolytic activities regarding ceramides, have been proposed for AdipoR2. Here, we have implemented MD analysis to further identify commonalities that may apply to the ligands screened in this study.
Overall, the discovery of novel compounds represents an important step in developing therapeutics toward the amelioration of pathologies associated with adiponectin and its receptors. Here, we have addressed this need through a computationally-aided examination as an initial platform for the characterization of the important chemical features of these receptors and their associated ligands.
2 Methods and materials
2.1 Protein preparation
The crystal structures of the adiponectin receptors were retrieved from the Protein Data Bank (www.rcsb.org) [40] for chains A, PDB ID: 5LXG [14] and 5LX9 [41] for AdipoR1 and AdipoR2 respectively. Both proteins were prepared using the protein Preparation Wizard protocol available in the Schrödinger suite (Maestro 10.4, Schrödinger, LLC) [42]. Briefly, hydrogens were added, bond orders were assigned, and loops and side chains were filled using the Prime module (Schrödinger, LLC) [43, 44]. Restrain minimization was performed using Optimized Potentials for Liquid Simulations (OPLS2005) force field until the RMSD reached 0.3 Å from the initial geometry in order to improve steric clashes [45, 46]. Additionally, PDB: 3WXV [3], A for AdipoR1 was used and prepared structurally as stated above.
2.2 Ligands retrieval and filtering
All of the ligands were retrieved by designing initial common scaffolds considering the proposed ceramidase activity in the presence of FFA in AdipoR2. Briefly, the ceramidase activity takes through the hydrolysis of the amide bond of the ceramide. Further details on these mechanisms can be found in Vasiliauskaité-Brooks et al. [14] regarding FFA interactions with AdipoR2, and in Airola et al. [23] for neutral ceramidase activity. Scaffolds were used to fit the geometry and volume adjacent to the zinc ion. These structures were used as queries in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) to retrieve ligands for docking. A manual search was also performed to retrieve additional compounds that include natural ligand as previously reported in the literature [47].
Collectively, 21,845 ligands were filtered by Lipinski’s rule of five (RO5), rapid elimination of swill (REOS) and pan assay interference compounds (PAINS 1, 2, and 3) using Canvas (Schrödinger, LLC). The resulting 20,067 (Data available at) ligands were used for molecular docking studies.
2.3 Binding pocket identification
2.3.1 Initial identification
The structures for AdipoR1 and AdipoR2 were prepared as previously described using Maestro Protein Wizard and then uploaded into SiteMap (Schrödinger, LLC). The software identified the top five possible binding sites and determined their potential druggability. Sites with a minimum of 15 points were identified with a more restrictive definition of hydrophobicity using a fine grid cropping at 4 Å (for display purposes only). SiteMap generates several physiochemical properties as described by Halgren [31, 33]. A SiteScore value of 1.0 and above is indicative of high druggability and this, in conjunction with Dscores, served as a measure of hydrophobicity [31, 33, 48]. SiteMap assigns a penalty to the high hydrophilic site that is reflected in the Dscore. Here, only the first site for both receptors was included for further analysis. Interestingly, the region generated by SiteMap fully accommodates the oleic acid present in the crystallographic structures for AdipoR2.
2.3.2 AdipoRs binding site further analysis
In order to further validate the binding region on the AdipoRs we applied IDD-based methodology [34]. Briefly, in IDD, the Gaussian network model generates frequency modes and the one that determines the highest degree of fluctuations is considered to be most associated with the biological activity of the protein. The determined slowest mode was then analyzed. The sign of the eigenvectors, associated with the residues, are considered as movements in positive and negative directions. Residues can be grouped by the same direction of motion and the two largest groups (domains) from the opposite directions are analyzed. The plane that best describes the separation between the two domains is defined as the D-plane. This plane is calculated by linear discriminant analysis, which determines the best separation axis (D-normal plane) between the two afore mentioned domains. Points that represent transitions between the domains (positive and negative direction of motion) are used for principal components analysis. Transition points are projected on the D-plane in order to define the D-axis. Catalytic sites have been described as a function of the IDD [34]. Moreover, the residues adjacent to the IDD have been shown to contain the majority of the possible catalytic sites for enzymatic activity [34]. More details regarding the algorithm can be found in the literature [34, 35, 49].
2.4 Docking
The binding site was assessed for docking of the ligands due to its large geometry and characteristics, as described above. The selection follows the notion that large cavities with high SiteScores may predict and accommodate ligands due to their large volume. This may also overcome the flexibility deficiency of the receptor during docking and present an opportunity for the initial assessment of the ligands in the receptor-ligand complex. The docking analysis relies on the creation of a receptor grid, which was calculated using Glide (Schrödinger, LLC). The grids for the two receptors were generated using Glide’s default settings. All of the ligands were prepared using LigPrep (Schrödinger, LLC) by identifying 32 stereoisomers with protonation states of pH 7 ± 2 using Epik (Schrödinger, LLC) using the OPLS2005 as a force field. The docking was generated initially using the standard precision (SP) protocol and the 10% ligands were further submitted for a second round of docking using the extra precision (XP) docking protocol in Glide. The top 1% of the ligands were used for molecular dynamics simulations.
Finally, a recent study hypothesized that byproducts of ceramidase activity may result in structural changes to the receptors [14]. Although this may be similar for both adiponectin receptors, only AdipoR1 has deposited crystallographic structures in closed and open conformations. The differences between the two structures reside in the relatively large shift of the transmembrane domain 5 (TM5) and intracellular loop 2. Therefore, we further analyzed the AdipoR1 in the closed conformation, PDB: 3WXV. The initial rigid docking produced very low Glide docking scores below − 7.3 kcal/mol (− 5 kcal/mol average). Therefore, we generated two different Glide’ grids: the first by rotation of the hydroxyl and thiol groups for residues: C183, S187, S219, S277, T282, and Y310; and the second by manual adjustments of the rotamers for R267, F271, and Y310. To further analyze the possibilities, an induced fit docking (IFD) protocol was applied for PDB 3WXV as described by Sherman [50]. In Maestro (Schrödinger, LLC), the parameters were set with a Van der Waals scaling of 0.5 for a standard protocol of up to 20 positions and the final re-docking was set for the SP protocol with the top 20 structures within 30 kcal/mol; all other parameters were left as indicated by the default mode.
2.5 Molecular dynamics simulation
Simulations for both receptors were implemented considering the top 1% of the best ligand positions generated by Glide. All of the simulations were carried out using the MD Desmond package [51]. The crystal structures used were PDB 5LXG and 5LX9 for AdipoR1 and AdipoR2 respectively. The receptor and ligand complexes were set up in an orthorhombic box using a buffer condition of 10 Å. The orientation of the membranes from the Orientation of Protein in Membranes (OPM) database [52]. Ions were neutralized for the system, salt was added at a concentration of 0.15 M NaCl, and OPLS 2005 was used for the force field [53]. Long-range electrostatic interactions were calculated with the Ewald method using a cut-off of 9 Å for Van der Waals and Coulomb interactions. The simulation was carried out in an isothermal, isobaric ensemble (NPT) with an initial temperature of 300 °K and 1 bar of pressure. The temperature followed the Nose–Hoover method and the pressure was controlled by the Martyna–Tobias–Klein protocol [41, 54, 55]. The simulation was set for 200 ns and trajectories were recorded every 100 ps. The default relaxation protocol for the system was used. Representative structures were extrapolated from the simulation at 0, 100, and 200 ns for graphical representations.
3 Results and discussion
3.1 Binding sites characterization and architecture
3.1.1 Binding sites localization
The two adiponectin receptors have a unique architecture that is poorly represented in other 7TM proteins. The central cavity, that spans almost the full length of the protein in both receptors, presents an opening located at the intracellular side as represented in Fig. 2a and b. The main differences between the two receptors (PDB: 5LXG and 5LX9) are the transmembrane openings due to the spatial rearrangement of the ICL2 and the shift of the TM5, as illustrated in Fig. 2a.
Adiponectin receptors structures. a Surface representation of the two AdipoRs. The two crystallographic structures for AdipoR1 are illustrated on the left-hand side where the TM5, TM6, and ICL2 are highlighted in the yellow transparent surface. b Opening between the TM5 and TM6 across AdipoRs. The positioning of TM5 and TM6 determines the overall area of the opening, illustrated in yellow
The Sitemap analysis, to determine possible binding sites, located the docking regions in the central cavity approximately within the opening in both receptors, as illustrated in Fig. 2b. The first possible binding site produced a SiteScore of 1.084 and 1.257 for AdipoR1 and AdipoR2, and a Dscore of 1.118 and 1.349 for AdipoR1 and AdipoR2 (see SI.1); these values are associated with high druggability [33]. These sites are located in the central cavity of both adiponectin receptors including the region adjacent to the zinc ion. Concerning the closed conformation of the AdipoR1 for PDB 3WXV, the SiteMap analysis produced high scores for the first detected site. This site scored 1.200 and 1.247 for SiteScores and Dscores respectively; however, this region is located away from the central cavity. The second site presents a better localization within the upper portion of the central cavity at the bilayer interface (SiteScore 1.188 and D-Score1.289). Nevertheless, the small volume does not seem suitable for ligand screening, generating unrealistic ligand positions and very low scores in the order of − 5 kcal/mol. Interestingly, the mere re-organization of the residues triad: R267, F271, and, particularly, Y310 produced better scores. The vertical continuity of the binding region gave accessibility to the middle and lower portion of the central cavity, fitting the ligands in a more probable region with improved binding energy scores.
3.1.2 Binding sites by intrinsic dynamic domain analysis
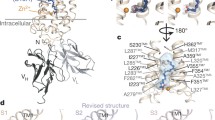
Further validation of the binding sites was provided by IDD. This type of inquiry has been used to determine catalytic sites in enzymes, suggesting that 90% of the sites can be found within 50% of the residues close to the D-plane [34]. As depicted in Fig. 3, the surface of the residues within 2 Å of the D-plane encapsulates the docked ligands. AdipoR1, in particular, contains a region that fully enclosed the most promising ligands. In Fig. 3, the frequency of residues is depicted by binning at 1 Å. The first bin contained R267, F271, and Y310, which may be key residues in modulation of the binding pockets in AdipoR1. In AdipoR2, the closest residue of the triad is R278 within 1 Å of the D-plane; the other two lie between 3 and 6 Å of the D-plane. As depicted in Fig. 3, the D-plane in AdipoR2 is located in the lower region of the central cavity in comparison to the AdipoR1 D-plane.
Intrinsic dynamic domains (IDD) of the AdipoRs. a AdipoR1, the left-hand side depicts the region covered by the closest residues to the D-plane. The top 1% of the screened ligands are shown. The right-hand side shows the residue frequencies binned at 1 Å, the graph below show the first bin within 1 Å from the D-plane. The triad of Arg 267, Phe 271, and Tyr 310 is within the 1 Å region. b AdipoR2 D-plane localization. The right-hand side, top graph illustrates the residues frequencies at 1 Å binning. The residues within 1 Å are further described in the bottom graph
3.1.3 Binding site architecture
The analysis of the binding region for the AdipoRs revealed differences in the geometry within the binding sites. As depicted in Fig. 4, the spatial orientation of the three key residues (AdipoR1: R267, F271, Y310 and AdipoR2:R278, F282, Y321) seems to be responsible for modulation in the architecture of the binding pockets of the two receptors. The residue Y310/321 seems to determine the extendibility of the binding region within the central cavity as illustrated in Fig. 4a and b.
AdipoRs binding cavity. a Binding site of the AdipoR1 with the residue triad delimiting the region. b AdipoR2 site with related residue delimiters of the region, note that the binding region (grey) is limited to the proximity of the ligand and, the entire cavity area is not depicted. c Overlay of the two binding regions onto the two AdipoRs. On top, the representation of the two receptors with their associated residues; bottom, top view through the cavity with the related residues
Site volume has been associated with pocket druggability, however, exact cut-off numbers vary by report and method [56,57,58]. In consideration of the foregoing the cavity volume was analyzed using KVFinder [24]. The volume of the region present in AdipoR1 was determined to be approximately 1000 Å3 and 900 Å3 for the region in AdipoR2. These values seem to be favorable for the druggability of the sites, as suggested by some reports [56,57,58]. The values of the volume alone may only further confirm the suitability of the binding site, rather than determine specific differences in the receptor binding characteristics.
Interestingly, the comparison between the open and closed conformations of the AdipoR1 provides useful clues regarding the key features of the receptor at its conformational endpoints. Due to the two different deposited crystallographic structures for AdipoR1, the possible binding region was analyzed by accounting for the open and closed conformations of this receptor.
In AdipoR1, the rearrangement of the three key residues determines the continuity and volume of the binding region (Fig. 5). Considering AdipoR1 in PDB 3WXV (closed conformation), the region is divided into three separate areas due to R267, F271, and Y310 spatial organization. As shown in Fig. 5a–c, the three residues in PDB 3WXV cut through the cavity, generating an unsuitable region for ligand docking. However, by introducing different rotational isomers for the residues triad, the continuity of the binding region could be restored, as illustrated in Fig. 5d. In this case, the binding scores could be improved and feasible positions could be generated by ligand screening. Importantly, the discontinuity and geometry of the region did not allow for determining feasible positions for the 3WXV structure during ligand screening. The screening scores could only be improved by the rearrangement of the three key residues. Interestingly, the initial docking using the 3WXV structure produced very low Glide docking scores of approximately − 5 kcal/mol on average, as illustrated in Fig. 5 for 3WXV. Therefore, a second grid was generated by rotating the hydroxyl and thiol groups for residues: C183, S187, S219, S277, T282, and Y310. The second round of Glide docking showed better positioning of the ligand, as illustrated in SI.2b and f with improved Glide’s scores of − 8 kcal/mol. Moreover, the PDB 3WXV was further used for IFD, as described in the Sect. 2. Upon relaxation of the docking region (IFD protocol), the scores were further improved for the ligands. The binding pocket fully accommodated the ligands and retained integral verticality, as depicted in SI.2 a and e. At the present time, the only indication of activity inhibition is related to the mutation of the zinc catalytic site by H191 and H202 for AdipoR1 and AdipoR2 respectively in crude lysates [9, 14]. While computationally interesting, this observation warrants further experimental validation.
AdipoR1 binding pocket architecture. a Left-hand side PDB: 3WXV, cyan, fragmentation of the cavity is depicted. Right-hand side AdipoR1 open conformation and cavity arrangement. b Overlay of the two cavity conformations for the open and closed conformations, red residue refers to PDB: 3WXV. c Top view of the cavity from open conformation, red color residue for 3WXV. The residues in red are depicted to the left when they penetrate the central area of the binding cavity. Right-hand side: the residues in open conformation. d The closed conformation of 3WXV with a relaxation of R267, F271, and Y310. Here, the cavity can contain the ligands without being fragmented, as illustrated above
Finally, since no crystallographic structure for AdipoR2 in its open conformation is available, we can only speculate in support of the theory that both receptors may present similar modalities related to their ability to process substrates.
3.2 Ligand docking
The compounds retrieved from PubChem were docked for AdipoR1 (PDB: 5LXG) and AdipoR2 (PDB: 5LX9). The top 1% of the screened ligands presented docking scores between − 13.6 and − 16.4 kcal/mol for AdipoR1 and AdipoR2 respectively. A selection of the top compounds is reported in SI.3 and SI.4 for AdipoR1 and AdipoR2 (5LXG and 5LX9) respectively. In SI.3 and SI.4, the ligand index is correlated with the docking scores. From the initial 20,067 retrieved compounds, 200 ligands were retained after two rounds of docking in Glide SP and XP mode (see Sect. 2 for details). The majority were specific to the type of receptor; however, 46 ligands showed promiscuity between the AdipoRs. Regarding AdipoR1, the presence of an acetate or carbonate group at one end of the ligand seemed to produce higher docking scores. As reported in SI.3 for binding energies above − 12 kcal/mol, all of the structures contain terminal acetate/carbonate groups with very few exceptions. Also, the two oxygen atoms seem to contribute to contacts primarily with the zinc ions of the receptor. Modification of this group may be explored in further analyses. As illustrated in SI.5, the first protein–ligand complex is less exposed to the solvent compared to the other two complexes. In AdipoR2, the presence of acetate or carbonate did not influence the docking scores as can be deduced by analyzing the selection of ligands in SI. 4. All of the ligands, with few exceptions, have acetate, carbonate or similar groups across the range of energy scores presented in SI.4.
3.3 Molecular dynamics
3.3.1 MD analysis of AdipoR1
The root mean squared deviation (RMSD) and root mean squared fluctuation (RMSF) of the Cα for each ligand complex are shown in Fig. 6a and b for AdipoR1 and AdipoR2 respectively, note that the figure contains ligand indexes related to the receptor’s residue position.
The RMSD for the AdipoR1 simulation shows different variabilities across the ligands. If we consider the coefficient of variation at the Cα between the complexes for the entire duration of the simulation, variabilities of 10.3%, 6%, and 10.1% for Complex I, II and III respectively were determined. However, notice that Complex I shows equilibration after approximately 70 ns and stabilizes within a shift of ~ 1 Å for the remainder of the simulation; this constitutes the lowest variability of the three complexes. As depicted in Fig. 6a, the overall shift for Complex I and III is around 3 Å, which may describe large conformational changes in the protein considering the rearrangement of the binding pocket as depicted in SI.5. Moreover, the analysis of the RMSF can give specific clues about the nature of these shifts. The five peaks, 1–5 in Fig. 6b, refer to specific residues, which compose the extracellular loop 1 (ECL1), TM3, (ICL2), TM5, and ICL3 respectively. Large shifts are associated with ECL1 and ICL2 and, arguably, this could be due to the more flexible nature of the loop structures, even though ICL2 has shown a pronounced translocation between the open and closed conformations of AdipoR1 [14]. Concerning TM5 and its positional shift between the open and closed conformations, a change is present only with ligand one and three but not with ligand two. Representations of the spatial rearrangement of the binding pocket during MD simulation at 0, 100, and 200 ns within 5 Å of the ligand are illustrated in SI.5.
3.3.2 MD analysis of AdipoR2
The RMSD for the AdipoR2 simulation shows an overall uniformity across the three complexes, illustrated in Fig. 6c. The coefficient of variation for the 200 ns simulation varies from 9.1%, 7.2% and 11.7% for the three complexes. Analysis of the RMSF shows variation in TM5 and ECL1, but also Complex II shows a peak related to TM3 that, is absent in the other complexes and has a smaller peak for TM5. This may be related to the lesser exposure of the ligand to the solvent compared to other ligands, as depicted in the openings in SI.5 e (bottom left).
As illustrated in Fig. SI.5 d–f, the AdipoR2 produced a more compact structure, which was expected, in comparison to the crystallographic structure of AdipoR1 due to the shift in TM5 (PDB: 5LXG). All three complexes showed similarities regarding the openings in the bottom part of the receptor as depicted in SI.5d–f (bottom left). Although in Complex II the opening is absent at 200 ns of simulation. These openings are located at the bottom of the receptor toward the N-terminal and they were more accentuated with the first ligand–protein complex (SI.5a, bottom left). This may represent a different mechanism for the substrate released by AdipoR2 in comparison to AdipoR1, which presents a central cavity more open to the surrounding solvent. Cavity rearrangements are present in AdipoR2 but these are less pronounced than in AdipoR1.
3.3.3 Receptor-ligand complex interactions
The contacts and types of interactions in the ligand-receptor complexes are illustrated in SI.6 for AdipoR1 and SI.7 for AdipoR2. These figures contain representative snapshots of the simulation showing the key residues interacting for more than 30% of the 200 ns.
3.3.3.1 AdipoR1-ligand complexes
Regarding Complex I, in the first 30 ns of the simulation, initial interactions in the form of Π–Π stacking between Y209 and Y310 with the benzene and pyran rings of the ligand were established. The portion of the ligand between the benzene ring and the carbonate group is initially exposed to solvent. The carbonate group is in contact with the zinc by coordination. Water also establishes hydrogen bonds with the single-bond of the oxygen from the carbonate. Residues H191, H337, H341, and D208 interact directly with the zinc in the presence of a water molecule. In the next 70 ns, the ligand becomes more buried in the cavity and the interactions include only the carbonate group. Another water molecule and Y317 interacted with the single bonded oxygen of the carbonate. In the last time period, water molecules are not present, leaving the zinc and residue Y317 interacting directly with the ligand by hydrogen bonding. The overall location of the first ligand is buried in the receptor and less exposed to the solvent compared to the other two ligands. The zinc ion shift is pronounced (around 4 Å), as shown in SI.5a (final snapshot of the simulation). This may well reflect the opening and closing of the receptor upon binding, however, this warrants further analysis.
In Complex II, the ligand is more exposed to the solvent, as depicted in SI.5b and SI.6. The residue triad: H191, H337 and H341 with D208 are still present throughout the simulation, as described for Complex I above. In the first 30 ns, the oxygens of the acetate group contact the zinc ion by metal coordination. The benzene ring, at the opposite end of the ligand from the acetate group, is stabilized by Y310 Π–Π stacking. The Y209 interacts by Π-cation with diethyl(methyl)azanium, CC[NH +](C)CC group possibly subtracting the bond from the adjacent benzene ring. In the next 70 ns the zinc ion disengages from the double oxygen bond of the acetate group and a hydrogen bond is established with water in the presence of Y194. The last part of the simulation shows that Y209 establishes contact with a water molecule by binding with the oxygen present in the middle of the ligand. As depicted in Figs. SI.5b and SI.6, the ligand is exposed to the solvent throughout the simulation.
In Complex III, as described for the other two complexes the zinc ion is stabilized by the three histidine residues, however, D208 directly interacts with the ethyl(methyl)azanium, CC[NH2 +]C group possibly preventing direct interactions with the zinc. Interestingly, this ligand shares an identical scaffold with the second ligand. The only difference is the addition of propane to the ethyl(methyl)azanium. This small modification may explain the differences in the binding scores between these two ligands, particularly considering the rearrangement of D208 and the contact with the zinc ion.
3.3.3.2 AdipoR2-ligand complexes
In Complex I the repeating pattern of the three histidine residues (H202, H348, and H352 for AdipoR2) is present with D219 and water during the first 30 ns of the simulation. Y328 forms hydrogens bonds with the double-bonded oxygen, which interact with the three surrounding water molecules. These water molecules are involved through hydrogen bonds with R275, R278, and Y220. Between 30 and 100 ns the Y328 produces Π–Π stacking with the benzene ring close to the acetate group; this terminal part of the ligand is more exposed to the solvent. The last part of the simulation shows the zinc interacting with the single bonded oxygen. Residue D219 reforming hydrogen bonds with a water molecule and F351 generates Π–Π stacking with the benzene ring, as illustrated in the last frame of SI.7. As illustrated in SI.5, during simulation, the receptor has openings on its lower part near the C-terminal. These openings, illustrated in SI.5, are present for all of the complexes with varying size, whether these openings constitute conformational changes identified by the simulation cannot be confirmed at this point.
The second complex present the ligand fully enclosed in the cavity however, a transitory small opening exposes the single-bonded oxygen of the acetate group to a water molecule and D219 at the beginning of the simulation. Residue F351 initially stabilizes the benzene ring to the opposite side buried in the pocket. Here in contrast to the others ligand, S198 is present with metal coordination with the zinc ion. The same histidine triad (H202, H348, and H352) is also present. In the remaining time of the simulation (200 ns), the S198 also interacts with the single- bonded oxygen of the acetate group by hydrogen bond. This represents the most stable simulation in terms of residue interactions with the ligand.
Complex III also has the zinc ion bound to the histidine triad and two water molecules generating hydrogen bonds with the double-bonded oxygen urea close to the piperidine ring. Π–Π stacking was also present between the central benzene ring and F351. The rest of the simulation generated the displacement of water and direct hydrogen bond of the urea oxygen with R278. In Complex III, the ligand becomes increasingly exposed to the solvent during simulation, as in Complex I, with the terminal acetate group exposed to the solvent and to the zinc interaction.
3.3.4 Commonalities across the ligands and possible mechanisms of action
Although the ceramidase activity of these receptors is low, some reports have highlighted the possibility that this activity may play an important role in its overall effects on lipid and glucose regulation [9, 14, 22]. A possible mechanism of action for these receptors has been proposed with the zinc ion acting as a catalyst for the hydrolytic reaction with sphingosine and fatty acids [14, 23]. The details of this activity have been suggested in the literature, particularly in AdipoR2 [14]. At the time of writing, this is the only full conceptual model describing the ceramidase activity of AdipoR2. For this reason, we have evaluated and compared the simulation results with the aforementioned model and with other possibilities of zinc-dependent activities.
In general, the presence of the zinc ion and the coordination to histidine residues and aspartic acid have been described for several enzymatic activities in metalloproteases and amino acetylases in the presence of water [59, 60]. In metalloproteases, the presences of motifs containing glutamine-histidine or aspartic acid-histidine combinations are common, with the zinc ion directly interacting with the residues. The water molecule and zinc ion represent a nucleophile center for a general base reaction. This reaction occurs with specific residues’ motifs across different types of proteins [59]. A subclass of metallo enzymes known as zinc hydrolase has been shown to have different catalytic site composition in the hydrolysis of different substrates [59]. In AdipoR2, a possible mechanism of action regarding the receptor’s ceramidase activity has been proposed by Vasiliauskaité-Brooks et al. [14], suggesting that the H348 mediates the proton transfer in a nucleophilic reaction involving the attack of the carbonyl bond in order to cleave the amide bond of the FFA. Moreover, the same study does not eliminate the possibility that D219 in AdipoR2 may act similarly to H348 as a base for proton retrieval from water. Considering the simulation presented here with the screened ligands, generally, histidine and aspartic acid are present, stabilizing the zinc ion and interacting with the ligands in a similar fashion in all of the complexes. Importantly, D219 (AdipoR2) and D208 (AdipoR1) interact with the oxygens present in the ligand’s terminal group. Considering, the simulation the presence of water molecules close to the zinc ion may support the nucleophilic addition at the acetic acid/carbonate region of the ligands for both of the AdipoRs. Furthermore, it seems more likely that aspartic acid may mediate the reaction instead of histidine with these compounds. A single generalization for a possible mechanism of action for all of the ligands remains difficult.
4 Conclusions
The generation of suitable ligands that modulate the adiponectin receptors represents an important step in the development of promising therapeutics toward the amelioration of several metabolic physiopathologies. Recently, both receptors have been crystallized but little is known about their biochemical properties and associated ligands. Very little literature is available and, therefore, the identification of new ligands represents a crucial step toward understanding these proteins and their underlying biochemistry as related to different metabolic conditions.
In the present study, we apply a computational approach to determine the binding pocket location and druggability applicable to subsequent virtual screening. The molecular docking identified hits for both AdipoRs with high binding energy scores of − 13.6 kcal/mol and − 16.5 kcal/mol for AdipoR1 and AdipoR2 respectively with PDBs 5LXG and 5LX9.
Moreover, we examined the two crystal structures for AdipoR1 (3WXV and 5LXG) due to the hypothesized open and closed conformations of this receptor because two crystallized proteins exist in different conformations. The rearrangement of the residue triad (R278, F282, and Y310) in AdipoR1 revealed changes to the geometry of the binding pocket, thereby altering its vertical continuity, which may underlie the differences in the binding scores.
MD analysis showed similar characteristics with the proposed ceramidase activity, at least for AdipoR2. The presence of aspartic acid interacting with the ligand R-group may suggest that hydrolytic activity is possible but involves aspartic acid instead of histidine as a mediator of the reaction.
In conclusion, we have identified amenable compounds for the AdipoRs. These docked hits represent starting chemical structures for further biochemical analyses considering the limited literature on these newly identified receptors. Furthermore, we have started to address the need for novel compounds required for future research, which are necessary for the understanding of these proteins and their associated pathologies.
Availability of data and materials
The complete dataset of 20,067 ligands analyzed during the current study is available from the corresponding author on reasonable request. Additional data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Kadowaki T, Yamauchi T, Kubota N (2008) The physiological and pathophysiological role of adiponectin and adiponectin receptors in the peripheral tissues and CNS. FEBS Lett 582:74–80. https://doi.org/10.1016/j.febslet.2007.11.070
Kadowaki T, Yamauchi T (2005) Adiponectin and adiponectin receptors. Endocr Rev 26:439–451. https://doi.org/10.1210/er.2005-0005
Tanabe H, Fujii Y, Okada-Iwabu M, Iwabu M, Nakamura Y, Hosaka T, Motoyama K, Ikeda M, Wakiyama M, Terada T et al (2015) Crystal structures of the human adiponectin receptors. Nature 520:312–316. https://doi.org/10.1038/nature14301
Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M et al (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423:762–769. https://doi.org/10.1038/nature01705
Yamauchi T, Kadowaki T (2008) Physiological and pathophysiological roles of adiponectin and adiponectin receptors in the integrated regulation of metabolic and cardiovascular diseases. Int J Obes 32:S13–S18
Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J-I, Hotta K, Shimomura I, Nakamura T, Miyaoka K et al (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun 257:79–83. https://doi.org/10.1006/bbrc.1999.0255
Ouchi N, Parker JL, Lugus JJ, Walsh K (2011) Adipokines in inflammation and metabolic disease. Nat Rev Immunol 11:85–97. https://doi.org/10.1038/nri2921
Almabouada F, Diaz-Ruiz A, Rabanal-Ruiz Y, Peinado JR, Vazquez-Martinez R, Malagon MM (2013) Adiponectin receptors form homomers and heteromers exhibiting distinct ligand binding and intracellular signaling properties. J Biol Chem 288:3112–3125. https://doi.org/10.1074/jbc.M112.404624
Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM et al (2011) Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med 17:55–63
Otvos L, Knappe D, Hoffmann R, Kovalszky I, Olah J, Hewitson TD, Stawikowska R, Stawikowski M, Cudic P, Lin F et al (2014) Development of second generation peptides modulating cellular adiponectin receptor responses. Front Chem. https://doi.org/10.3389/fchem.2014.00093
Polito R, Nigro E, Pecoraro A, Monaco ML, Perna F, Sanduzzi A, Genovese A, Spadaro G, Daniele A (2019) Adiponectin receptors and pro-inflammatory cytokines are modulated in common variable immunodeficiency patients: correlation with Ig replacement therapy. Front Immunol. https://doi.org/10.3389/fimmu.2019.02812
Kelesidis I, Kelesidis T, Mantzoros CS (2006) Adiponectin and cancer: a systematic review. Br J Cancer 94:1221–1225. https://doi.org/10.1038/sj.bjc.6603051
Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N et al (2001) The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 7:941–946
Vasiliauskaité-Brooks I, Sounier R, Rochaix P, Bellot G, Fortier M, Hoh F, De Colibus L, Bechara C, Saied EM, Arenz C et al (2017) Structural insights into adiponectin receptors suggest ceramidase activity. Nature. https://doi.org/10.1038/nature21714
Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, Funk WD (2005) PAQR proteins: a novel membrane receptor family defined by an Ancient7-transmembrane pass motif. J Mol Evol 61:372–380. https://doi.org/10.1007/s00239-004-0375-2
Wess J (1997) G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J 11:346–354
Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, Iwabu M, Okada-Iwabu M, Kawamoto S, Kubota N, Kubota T et al (2007) Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13:332–339
Heiker JT, Wottawah CM, Juhl C, Kosel D, Mörl K, Beck-Sickinger AG (2009) Protein kinase CK2 interacts with adiponectin receptor 1 and participates in adiponectin signaling. Cell Signal 21:936–942. https://doi.org/10.1016/j.cellsig.2009.02.003
Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M et al (2010) Adiponectin and AdipoR1 regulate PGC-1[agr] and mitochondria by Ca2 + and AMPK/SIRT1. Nature 464:1313–1319
Lee M-H, Klein RL, El-Shewy HM, Luttrell DK, Luttrell LM (2008) The adiponectin receptors AdipoR1 and AdipoR2 activate ERK1/2 through a Src/Ras-dependent pathway and stimulate cell growth. Biochemistry 47:11682–11692. https://doi.org/10.1021/bi801451f
Zhou L, Deepa SS, Etzler JC, Ryu J, Mao X, Fang Q, Liu DD, Torres JM, Jia W, Lechleiter JD et al (2009) Adiponectin activates AMP-activated protein kinase in muscle cells via APPL1/LKB1-dependent and phospholipase C/Ca2 +/Ca2 +/calmodulin-dependent protein kinase kinase-dependent pathways. J Biol Chem 284:22426–22435. https://doi.org/10.1074/jbc.M109.028357
Holland WL, Xia JY, Johnson JA, Sun K, Pearson MJ, Sharma AX, Quittner-Strom E, Tippetts TS, Gordillo R, Scherer PE (2017) Inducible overexpression of adiponectin receptors highlight the roles of adiponectin-induced ceramidase signaling in lipid and glucose homeostasis. Mol Metab 6:267–275. https://doi.org/10.1016/j.molmet.2017.01.002)
Airola MV, Allen WJ, Pulkoski-Gross MJ, Obeid LM, Rizzo RC, Hannun YA (2015) Structural basis for ceramide recognition and hydrolysis by human neutral ceramidase. Structure 23(8):1482–1491
Oliveira SH, Ferraz FA, Honorato RV, Xavier-Neto J, Sobreira TJ, de Oliveira PS (2014) KVFinder: steered identification of protein cavities as a PyMOL plugin. BMC Bioinform 15:197. https://doi.org/10.1186/1471-2105-15-197
Bromhead C, Miller JH, McDonald FJ (2006) Regulation of T-cadherin by hormones, glucocorticoid and EGF. Gene 374:58–67. https://doi.org/10.1016/j.gene.2006.01.013
Hug C, Wang J, Ahmad NS, Bogan JS, Tsao T-S, Lodish HF (2004) T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci USA 101:10308–10313. https://doi.org/10.1073/pnas.0403382101
Hebbard LW, Garlatti M, Young LJ, Cardiff RD, Oshima RG, Ranscht B (2008) T-cadherin supports angiogenesis and adiponectin association with the vasculature in a mouse mammary tumor model. Can Res 68:1407–1416. https://doi.org/10.1158/0008-5472.can-07-2953
Ruan H, Dong LQ (2016) Adiponectin signaling and function in insulin target tissues. J Mol Cell Biol 8:101–109. https://doi.org/10.1093/jmcb/mjw014
Okada-Iwabu M, Yamauchi T, Iwabu M, Honma T, Hamagami K-I, Matsuda K, Yamaguchi M, Tanabe H, Kimura-Someya T, Shirouzu M et al (2013) A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 503:493–499. https://doi.org/10.1038/nature12656
Ma B, Shatsky M, Wolfson HJ, Nussinov R (2002) Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci Publ Protein Soc 11:184–197
Halgren T (2007) New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 69:146–148. https://doi.org/10.1111/j.1747-0285.2007.00483.x
Sirin S, Kumar R, Martinez C, Karmilowicz MJ, Ghosh P, Abramov YA, Martin V, Sherman W (2014) A computational approach to enzyme design: predicting ω-aminotransferase catalytic activity using docking and MM-GBSA scoring. J Chem Inf Model 54:2334–2346. https://doi.org/10.1021/ci5002185
Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49:377–389. https://doi.org/10.1021/ci800324m
Li H, Sakuraba S, Chandrasekaran A, Yang L-W (2014) Molecular binding sites are located near the interface of intrinsic dynamics domains (IDDs). J Chem Inf Model 54:2275–2285. https://doi.org/10.1021/ci500261z
Chandrasekaran A, Chan J, Lim C, Yang L-W (2016) Protein dynamics and contact topology reveal protein–DNA binding orientation. J Chem Theory Comput 12:5269–5277. https://doi.org/10.1021/acs.jctc.6b00688
Yang L-W, Rader AJ, Liu X, Jursa CJ, Chen SC, Karimi HA, Bahar I (2006) oGNM: online computation of structural dynamics using the Gaussian Network Model. Nucleic Acids Res 34:W24–W31. https://doi.org/10.1093/nar/gkl084
Li H, Chang Y-Y, Lee JY, Bahar I, Yang L-W (2017) DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res 45:W374–W380. https://doi.org/10.1093/nar/gkx385
Ertl P, Jelfs S, Mühlbacher J, Schuffenhauer A, Selzer P (2006) Quest for the rings. in silico exploration of ring universe to identify novel bioactive heteroaromatic scaffolds. J Med Chem 49:4568–4573. https://doi.org/10.1021/jm060217p
Lipinski C, Hopkins A (2004) Navigating chemical space for biology and medicine. Nature 432:855–861
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101:4177–4189. https://doi.org/10.1063/1.467468
Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins Struct Funct Bioinform 55:351–367. https://doi.org/10.1002/prot.10613
Jacobson MP, Friesner RA, Xiang Z, Honig B (2002) On the Role of the crystal environment in determining protein side-chain conformations. J Mol Biol 320:597–608. https://doi.org/10.1016/S0022-2836(02)00470-9)
Jorgensen WL, Tirado-Rives J (2005) Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci USA 102:6665–6670. https://doi.org/10.1073/pnas.0408037102
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
Sun Y, Zang Z, Zhong L, Wu M, Su Q, Gao X, Zan W, Lin D, Zhao Y, Zhang Z (2013) Identification of adiponectin receptor agonist utilizing a fluorescence polarization based high throughput assay. PLoS ONE 8:e63354. https://doi.org/10.1371/journal.pone.0063354
Vidler LR, Brown N, Knapp S, Hoelder S (2012) Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem 55:7346–7359. https://doi.org/10.1021/jm300346w
Bartlett GJ, Porter CT, Borkakoti N, Thornton JM (2002) Analysis of catalytic residues in enzyme active sites. J Mol Biol 324:105–121. https://doi.org/10.1016/S0022-2836(02)01036-7)
Sherman W, Day T, Jacobson MP, Friesner RA, Farid R (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. https://doi.org/10.1021/jm050540c
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, et al (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE conference on Supercomputing, Tampa, Florida, ACM, p 84
Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI (2006) OPM: orientations of proteins in membranes database. Bioinform (Oxford, England) 22:623–625. https://doi.org/10.1093/bioinformatics/btk023
Ahmed A, Villinger S, Gohlke H (2010) Large-scale comparison of protein essential dynamics from molecular dynamics simulations and coarse-grained normal mode analyses. Proteins 78:3341–3352. https://doi.org/10.1002/prot.22841
Nosé S (1984) A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys 81:511–519. https://doi.org/10.1063/1.447334
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31:1695–1697
Pérot S, Sperandio O, Miteva MA, Camproux A-C, Villoutreix BO (2010) Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discov Today 15:656–667. https://doi.org/10.1016/j.drudis.2010.05.015)
Nayal M, Honig B (2006) On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins 63:892–906. https://doi.org/10.1002/prot.20897
An J, Totrov M, Abagyan R (2005) Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics MCP 4:752–761. https://doi.org/10.1074/mcp.M400159-MCP200
Hernick M, Fierke CA (2005) Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch Biochem Biophys 433:71–84. https://doi.org/10.1016/j.abb.2004.08.006)
McCall KA, Huang C, Fierke CA (2000) Function and mechanism of zinc metalloenzymes. J Nutr 130:1437s–1446s. https://doi.org/10.1093/jn/130.5.1437S
Acknowledgements
Open access funding provided by University of Gothenburg. The authors want to thank Prof. Patrik Rorsman for helpful discussions and support.
Funding
This work was funded by the Swedish Medical Research Council (Grant ID: FE-57 2021005208) and Wilhelm & Martina Lundgrens Vetenskapsfond (2016-1266).
Author information
Authors and Affiliations
Contributions
MM conceived and designed the study, carried out the simulations, analyzed the data and drafted the manuscript. AMK critically revised the manuscript and helped in draft the manuscript. All authors gave final approval for publication.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muratore, M., Komai, A.M. Theoretical study of the adiponectin receptors: binding site characterization and molecular dynamics of possible ligands for drug design. SN Appl. Sci. 2, 533 (2020). https://doi.org/10.1007/s42452-020-2333-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2333-z