Abstract
Alpha-glucosidases are involved in the hydrolyzation of glycosidic bond of di- or oligo-saccharides into mono-saccharides, thus help in the breakdown and absorption of sugars. Inhibition of alpha-glucosidases by inhibitors tend to slow break down and release of sugars into the bloodstream and can be used as therapeutic agents in the treatment of diabetes and obesity. In particular, some of the inhibitors are used in the treatment mainly acarbose, voglibose, and miglitol. In this study, we have reported the alpha-glucosidase inhibitory compound from a newly described marine bacterium Arthrobacter enclensis. The purified compound from A. enclensis was identified by HPLC, and further FTIR and tandem mass spectrometry (MS/MS) methods. The purified compound was annotated and identified by a Web tool CFM-ID (Competitive Fragmentation Modeling for Metabolite Identification). From analysis it was found that the compound showed high similarity with acarbose which is a C7N aminocyclitol compound. Further, we analyze the draft genome of A. enclensis using anti-SMASH. We observed that it matches the homology with biosynthetic gene cluster of acarviostatin and acarbose with 11% and 7% respectively. We demonstrated that, the purified compound from A. enclensis shows the inhibitory activity against alpha-glucosidase with an IC50 value of 500 ± 0.142 μg/ml as compared to standard acarbose, which showed an IC50 value of 200 ± 0.012 μg/ml. This result suggests that A. enclensis has a tendency to produce a C7N aminocyclitol like molecule which matched to acarbose and it uses different biosynthetic gene cluster for the synthesis of C7N aminocyclitol like molecule, which can be further used for the production and treatment purpose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Alpha-glucosidase (EC. 3.2.1.20) is the enzyme of glycoside hydrolase family which hydrolyzes the alpha-glycosidic bonds present in the carbohydrates. It cleaves the α-1, 4 linked terminal non-reducing end of a sugar moiety [10]. It helps in the breakdown of carbohydrates. Inhibition of alpha-glucosidase helps in slow release of monosaccharides into the bloodstream after a meal. In diabetes mellitus type 2 hyperglycemia conditions occurs after the meal due to the action of insulin resistance. To control the hyperglycemic condition after the meal, alpha-glucosidase inhibitors are used as therapeutic agents [1, 20].
Inhibitors of alpha-glucosidase are naturally produced by many organisms and several inhibitors have been reported till date, some are being already used for the treatment purpose. Many plants and microorganisms have been reported to produce glycosidases inhibitors, but those which produce compounds that mimick the structure of monosaccharide or oligosaccharide are very efficient because of their resemblance to the sugar moeity of natural substrate. C7N aminocyclitol are metabolites which contains oligosaccharides with amine derivatives. These compounds have a pseudosaccharide cyclitol core unit to which different numbers of sugars molecules are attached [12]. An example of this is Acarbose, which is an inhibitor for alpha-glucosidase which comprises an acarviosine moiety and maltose unit. Acarviosine is composed of an unsaturated cyclitol (valienol or valienamine) ring which is attached to the 4-amino-4,6-dideoxyglucose unit with an amide linkage [23]. This amino-oligosaccharide family contains other compounds as well which showed the inhibitory activity against glycosidase enzymes. This class of compounds is isolated from the Actinoplanes sp. [21] and some Streptomyces sp. [7, 14].
The genus Arthrobacter was described by Conn and Dimmick [4] for aerobic, Gram positives, rod shaped (young culture) to coccus shaped (in older cultures) bacteria. It had been demonstrated that, species of Arthrobacter were able to degrade hazardous contaminants and chemicals [6, 9]. Some Arthrobacter species have been reported to lyse yeast cells [11] and mycelium of Fusarium roseum [15, 19]. Despite of above activities, inhibition of alpha-glucosidase enzymes was not reported from any Arthrobacter species till date.
The aim of the study was to isolate alpha-glucosidase inhibitor from a novel marine bacterium Arthrobacter enclensis, which was previously, reported [5]. We had investigated its crude complex inhibitory activity against alpha-glucosidase. We have isolated and identified the compound that showed similarity with an acarbose, with a potent alpha-glucosidase inhibition activity.
2 Materials and methods
2.1 Microorganism and enzyme
Arthrobacter enclensis sp. nov., which was isolated from marine sediment sample from Chorao Island, Goa, India, was used in this study. Arthrobacter enclensis was stored at -80 °C in 20% (v/v) sterile glycerol until further use. Prior to the study, A. enclensis was activated on the nutrient agar plate at 30 °C for about 24 h. The alpha-glucosidase enzyme from Sac-charomyces cerevisiae (E.C.3.2.1.20), p-nitrophenyl-α-D-glucopyranoside (pNPG) and acarbose was purchased from Sigma (Merck). Other chemicals used were of analytical grade.
2.2 Fermentation of Arthrobacter enclensis
The seed culture was prepared by shaking at 30 °C for 24 h in a medium consisting of (g/L): glucose 15.0; peptone 7.5; K2HPO4·3H2O 1.0 and NaCl 5.0. The seed culture was then transferred to the fermentation medium consisting of (g/L): glucose-30.0; maltose-15.0; peptone-5.0; monosodium glutamate-3.0; K2HPO4·3H2O-1.0; MgSO4·7H2O-1.0; FeSO4·7H2O-0.02 and NaNO3-3.0. The fermentation was carried out in shaking condition at 30 °C for 168 h (7 days) at 140 rpm. The growth parameter of A. enclensis and the effect of their crude fermentation broth on alpha-glucosidase were monitored for 168 h (7 days) with an time interval of 24 h. Growth was measured as an optical density at spectrophotometer with wavelength of 600 nm and the inhibition were measured as a % inhibition.
2.3 Purification of alpha-glucosidase inhibitor
The culture broth was centrifuged at 8000 rpm for 10 min. The filtrate was then incubated with S. cerevisiae beads for 24 h at 37 °C. The S. cerevisiae beads were prepared according to the method described by [22]. The S. cerevisiae beads were removed by centrifugation at 6000 rpm for 10 min. The resulting supernatant was mixed with an equal volume of absolute cold ethanol and kept for 30 min to remove any soluble dextran. The mixture was centrifuged again and filtered through Whatman filter paper to remove the precipitated material.
The water fraction was then subjected to the gel permeation chromatography. The fraction was loaded on Biogel P-2 (extra fine) column (1.5 × 90 cm) and eluted with deionized water at a flow rate of 0.1 ml/min, to collect 10 ml fractions. The fractions which showed the inhibitory activity against alpha-glucosidase were pooled together and concentrated for further analysis.
2.4 Assay for alpha-glucosidase inhibition
The alpha-glucosidase inhibition assay was carried out at a 96 well microtiter plate based spectrometry (Thermo Multiskan EX). A volume of 50 μl sample and 100 μl of enzyme solution (1U/ml) prepared in 0.1 M sodium phosphate buffer of pH 6.9, were incubated in microtiter plates for 10 min at 25 °C. After incubation, 50 μl (5.0 mM) of para-nitrophenyl alpha-D-glucopyranoside (pNPG) solution prepared in 0.1 M sodium phosphate buffer of pH 6.9, was added in each well and further incubated for 5 min at 25 °C. The absorbance was recorded at 405 nm by using microtiter plate reader. The absorbance of samples was compared with the absorbance of control, which contains 50 μl of 0.1 M phosphate buffer of pH 6.9 in place of sample. The enzyme inhibition was expressed as % inhibition using the following equation.
2.5 High performance liquid chromatography (HPLC) analyses
The fractions collected from Biogel P-2 chromatography were further analyzed by HPLC along with standard acarbose. The samples were injected into HPLC system (Thermo, UHPLC Ultimate 3000); Carbohydrate column (Waters) 3.9 × 300 mm, 10 μl of injection volume was employed for HPLC analysis with a flow rate of 1 ml/min and a wavelength at 210 nm. The mobile phase used was acetonitrile: phosphate buffer (0.6 g of KH2PO4 and 0.48 g of Na2HPO4) with a ratio of 70:30.
2.6 Characterization of the alpha-glucosidase inhibitor
The isolated compound was identified using FTIR spectrometry, and by tandem mass spectrometry and compared with standard acarbose. The FTIR was investigated on BRUKER- Tensor37 system with a wavelength range from 4000 to 500 cm−1. The system was controlled by OPUS v.6.5 software. Mass detection (MS) and tandem mass spectro-metry (MS/MS) spectra were performed on a Thermo Q-Exactive Orbitrap mass spec-trometer, equipped with an ESI source and an ion trap mass analyzer. The mobile phase methanol: water (90:10) with 0.1% formic acid was used at a flow rate of 0.350 ml/min. The injection volume of 1.5 μl was used for positive mode ionization. The mass range of m/z is from 100 to 1500. For MS/MS analysis the collision energy was set at 35 V and system software was controlled by X-calibur v.3.0.
2.7 Anti-SMASH analysis of whole genome of A. enclensis
The draft genome of A. enclensis was generated using Illumina sequencing at DOE Joint Genome Institute (JGI), Walnut Creek, California, USA. By using Illumina HiSeq2000 platform, shotgun library and sequencing were constructed which generates 8,861,546 reads of totalling 1338.1 Mb data. The final assembly contained 19 contings in 18 scaf-folds. The length of circular map genome consists of 4,226,231 bp with G + C content of 67.08% (Fig. 1). The general features of draft genome statistics are summarized in (Table 1). This draft genome sequence of A. enclensis was deposited in DDBJ/ENA/GenBank under the accession number LNQM00000000 [16]. This draft genome of A.enclensis was analyzed using anti-SMASH (Antibiotics and Secondary Metabolite Analysis Shell) v.4.0.
Graphical circular map of A. enclensis genome. This genome consists of 4,226,231 bp and 3888 predicted protein coding sequence. From the outside to the center: protein coding sequences (blue colour), RNA genes (tRNA red, rRNA pink and others grey) on the forward strand, G+C content (black colour) and GC skew (purple negative value and olive positive value)
3 Results and discussion
3.1 Effect of crude fermentation broth on alpha-glucosidase and growth parameters of A. enclensis
Production of alpha-glucosidase inhibitors has gained large commercial interest due to its beneficial properties in the human health. The alpha-glucosidase inhibitory complex was isolated from the culture broth of A. enclensis. The starting inhibitory activity was observed from 24 h of fermentation which was then exceeded from 96 h to the 168 h. The growth pattern of A. enclensis was measured as an optical density with a wavelenght of 600 nm. The maximum growth of an organism was observed at 96 h of fermentation on which 50% of inhibition was achieved. The decline of growth was observed from 120 h of fermentation and effect of inhibition was seemed to be constant to the rest of days. The corelation between growth of bacteria to the inhibition against an enzyme is shown in (Fig. 2). It was then purified by Biogel P-2 gel permeation chromatography. The fractions from 8 to 12 showed the inhibition and was then pooled and further analyzed with HPLC, FTIR and MS/MS.
3.2 HPLC analysis of purified compound
The HPLC chromatogram of standard acarbose and purified compound of A. enclensis and unfermented broth as a control is shown in (Fig. 3). The standard acarbose showed a peak at a retention time of 6.61 min. The unfermented broth shows peaks mostly at ranging 2.0–4.4 min, while purified compound showed peaks at different retention times apart from 2.0 to 4.4 min, suggesting that some new peaks appeared which were not observed in unfermented broth. The purified compound showed a peak at a retention time 6.28 min. Apart from this, some additional peaks were also observed which might be of other components produced by A. enclensis.
3.3 IR of purified compound
The IR spectra of standard acarbose and purified compound of A. enclensis is shown in Fig. 4. The IR spectrum of acarbose displayed strong OH stretching (optimal at 3290 cm−1) at wide peak ranging from 3000 to 3800 cm−1. The C-H stretching appeared at 2875 cm−1. The C=C weak stretching has appeared at a wide range of 1420–1650 cm−1. The medium C–H bending and C–O stretching was displayed at 1364 and 1034 cm−1, respectively. The IR spectrum of a purified compound of A. enclensis showed a strong OH stretching at 3272 cm−1. The C–H stretching appeared at 2925 cm−1, C=C stretching at 1639 cm−1 and C–H bending at 1359 cm−1 ranges of a peak. The C–O stretching was displayed at 1012–1144 cm−1. From the above, it was found that most of the groups displayed correspond to the oligosaccharides. Acarbose is a pseudo-oligosaccharide compound, which contains N–H amide linkage, which usually ranges from 3360 to 3310 cm−1, and in case of acarbose it is occupied by a wide range of OH groups [18]. The IR spectra of the purified compound of A. enclensis also showed the presence of an oligosaccharide compound.
3.4 Mass spectrometry (MS) and tandem mass spectrometry (MS/MS)
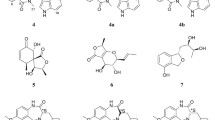
The purified compound was then identified by positive ion mode electrospray ionization mass spectrometry (ESI–MS) and tandem mass spectrometry (MS/MS) analysis. The ESI–MS of acarbose was done which showed a peak at m/z 646.2542 [M + H]+ signal is shown in (Fig. 5a). In full scan of an active compound of A. enclensis showed a peak at m/z 646.2545 [M + H]+ signal is shown in (Fig. 5b). The tandem mass spectrometry (MS/MS) spectrum of parental compounds of both standard acarbose and peak of m/z 646.2545 of A. enclensis was done (Fig. 5c, d). The fragmentation pattern of both the parental compounds was found to be similar. The annotation of peaks in the spectrum and the integration of spectrum were done by using a web tool CFM-ID. The CFM-ID provides the method for the efficiently identified metabolites in a spectrum for a known chemical structure generated by MS/MS [2]. The signals at 466 m/z and 304 m/z correspond to the loss of one to two glucose units from the molecule. This indicates the dissociation of glycosidic bond. The fragment ion 304 m/z, 268 m/z and 286 m/z were matched with acarviosine unit. The fragment 146 m/z indicates the cleavage at N–H (amide) linkage of acarviosine moiety. The 146 and 128 fragment ions were matched with cyclitol ring of an acarviosine molecule. From the above, it was observed that the fragments produced upon ionization were of dissociation products from a glycosidic bond and from an amide bond. By comparison of the spectrum of acarbose to the active compound of A. enclensis, it was matched highly with the acarbose. From spectrum analysis it was observed that the purified compound showed the matches with amide linkage of acarviosine moeity and glycosidic linkage to one glucose moeity, which is a characteristic of C7N aminocyclitol class compounds.
3.5 Alpha-glucosidase inhibition
We investigated the inhibitory effect of purified compound from A. enclensis and standard acarbose on alpha-glucosidase (Table 2). We found that standard acarbose showed the inhibitory activity with an IC50 value of 200 ± 0.012 μg/ml while the purified compound from A. enclensis showed the activity at an IC50 value of 500 ± 0.142 μg/ml. The maximum inhibition was observed 91.11% for acarbose and 75.42% for compound isolated from A. enclensis.
3.6 Anti-SMASH analysis of draft genome of A. enclensis
The anti-SMASH helps in the alignment of a query gene cluster with the known gene cluster from a database and to identify biosynthetic gene cluster [3, 13, 16]. We observed that the cluster 21 from scaffold 00004 from gene SMCF_980 showed the putative gene cluster similarity with acarviostatin biosynthetic gene cluster (11%) and acarbose bio-synthetic gene cluster (7%) from the location of 48801–67650 nucleotides with Cluster-Finder probability of 0.8674. In both gene clusters, it matches with acarbose 4-alpha-glucanotransferase (AcbQ/SctQ) and alpha-amylase (AcbZ/SctZ1 and SctZ2) genes (Fig. 6). The acarviostatin and acarbose both is C7N aminocyclitol compound and acarviostatin act as alpha-amylase inhibitor and acarbose as alpha-glucosidase and alpha-amylase inhibitor. The putative gene cluster of acarviostatin biosynthesis (Sct) exhibited similarity with gac-gene cluster of Streptomyces glaucescens GLA.O (GenBank accession AM409314, [17]) and with acb-gene cluster of Actinoplanes sp. SE50/110 (GenBank accession Y18523, [21]) for acarbose synthesis. Some steps of the putative intracellular synthetic pathway of acarviostatin are similar to the biosynthetic pathway of acarbose [8], suggesting that the formation of core cyclitol structure of both acarviostatin and acarbose shares some common steps.
Later on, we compare the protein encoding similarity of gene cluster of A. enclensis with acb-gene cluster of Actinoplanes sp. SE50/110 with reference gene cluster of S. glaucescens (Table 3). We found that, some putative gene products of A. enclensis showed similarity with acarbose gene products of Actinoplanes sp. SE50/110. We observed that alpha-amylase matches 54% with AcbZ, ABC transporter ATP binding protein matches 32% with AcbW, 4-alpha-glucanotransferase matches 46% with AcbQ, dTDP-glucose 4,6-dehydratase matches 44% with AcbB, alpha-amylase matches 48% with AcbE and ABC transporter permease and ABC transporter binding protein matches 55%, 54% and 45% with AcbG, AcbF and AcbH respectively.
4 Conclusion
A alpha-glucosidase inhibitory compound was isolated and purified from a novel marine bacteria A. enclensis. Inhibitory compound is identified based on HPLC, FTIR and tandem mass spectrometry (MS/MS) analysis. It was observation that, isolated compound showed a similarity with a C7N aminocyclitol like molecule. Further, we found that inhibitory compound showed the presence of acarviosine like ring in the compound from the MS/MS analysis and was annotated and confirmed by CFM-ID, and the purified compound of A. enclensis matches with the acarbose molecule. The draft genome of A. enclensis showed the biosynthetic gene cluster similarity with acarviostatin and acarbose. Both acarviostatin and acarbose are C7N aminocyclitol compounds and are known to possess glycosidases inhibition activities. From present study, we concluded that the A. enclensis has a tendency to produce C7N aminocyclitol like molecule which showed inhibitory activity towards alpha-glucosidase enzyme and matches high similarity with acarbose. This is the first time, we are reporting the alpha-glucosidase inhibitory compound from a newly described marine bacterium A. enclensis.
References
Alagesan K, Thennarasu P, Kumar V, Sankarnaraynan S, Balsamy T (2012) Identification of α-glucosidase inhibitors from Psidium guajava leaves and Syzygium cumini Linn. Seeds. Int J Pharm Sci Res 3:316–322
Allen F, Pon A, Wilson M, Greiner R, Wishart D (2014) CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res 42(W1):W94–W99
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de Los Santos E, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(W1):W36–W41
Conn HJ, Dimmick I (1947) Soil bacteria similar in morphology to Mycobacterium and Corynebacterium. J Bacteriol 54:291–303
Dastager SG, Qin L, Tang SK, Krishnamurthi S, Lee JC, Li WJ (2014) Arthrobacter enclensis sp. nov., isolated from sediment sample. Arch Microbiol 196:775–782
Fukatsu H, Goda M, Hashimoto Y, Higashibata H, Kobayashi M (2005) Optimum culture conditions for the production of N-substituted formamide Deformylase by Arthrobacter pascens F164. Biosci Biotechnol Biochem 69:228–230
Geng P, Sun T, Zhong Q, Li X, Shi L, Bai F, Bai G (2013) Two novel potent α-amylase inhibitors from the family of acarviostatins isolated from the culture of Streptomyces coelicoflavus ZG0656. Chem Biodivers 10:452–459
Guo X, Geng P, Bai F, Bai G, Sun T, Li X, Shi L, Zhong Q (2012) Draft genome sequence of Streptomyces coelicoflavus ZG0656 reveals the putative biosynthetic gene cluster of acarviostatin family α-amylase inhibitors. Lett Appl Microbiol 55:162–169
Jussila MM, Jurgens G, Lindström K, Suominen L (2006) Genetic diversity of culturable bacteria in oil-contaminated rhizosphere of Galega orientalis. Environ Pollut 139:244–257
Kimura A, Lee J, Lee I, Lee H, Park K, Chiba S, Kim D (2004) Two potent competitive inhibitors discriminating α-glucosidase family I from family II. Carbohydr Res 339:1035–1040
Kitamura K, Kaneko T, Yamamoto Y (1972) Lysis of viable yeast cells by enzymes of Arthrobacter luteus. J Gen Appl Microbiol 18:57–71
Mahmud T (2003) The C7N aminocyclitol family of natural products. Nat Prod Rep 20(137):166
Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39(Web Server issue):W339–W346
Meng P, Xie C, Geng P, Qi X, Zheng F, Bai F (2013) Inhibitory effect of components from Streptomyces species on α-glucosidase and α-amilase of different origin. Appl Biochem Microbiol 49:160–168
Morrissey R, Dugan E, Koths J (1976) Chitinase production by an Arthrobacter sp. lysing cells of Fusarium roseum. Soil Biol Biochem 8:23–28
Neurgaonkar PS, Dharne MS, Dastager SG (2016) Draft genome sequence of Arthrobacter enclensis NCIM 5488T for secondary metabolism. Genome Announcements. https://doi.org/10.1128/genomea.00497-16
Rockser Y, Wehmeier U (2009) The gac-gene cluster for the production of acarbose from Streptomyces glaucescens GLA.O-identification, isolation and characterization. J Biotechnol 140:114–123
Saoud A, Akowuah GA, Fatokun O, Mariam A, Khalivulla SI (2017) Determination od Acarbose in tablets by attenuated total reflectance Fourier transform infrared spectroscopy. Arch Ind Biotechnol 1:20–26
Szajer C, Koths JS (1973) Physiological properties and enzymatic activity of an Arthrobacter capable of lysing Fusarium sp. Acta Microbiol 5:81–86
Wang L, Hou Y, Peng J, Qi X, Zhang Q, Bai F, Bai G (2012) Bioactivity -based HPLC Tandem Q/TOF for alpha glucosidase inhibitors. Screening, identification and quantification from Actinomycetes. Lat Am J Pharm 31:693–698
Wehmeier UF, Piepersberg W (2004) Biotechnology and molecular biology of the α-glucosidase inhibitor acarbose. Appl Microbiol Biotechnol 63:613–625
Yoon S, Mukerjea R, Robyt JF (2003) Specificity of yeast (Saccharomyces cerevisiae) in removing carbohydrates by fermentation. Carbohydr Res 338:1127–1132
Zhang CS, Stratmann A, Block O, Bruckner R, Podeschwa M, Altenbach HJ, Wehmeier UF, Piepersberg W (2002) Biosynthesis of the C7-cyclitol moiety of acarbose in Actinoplanes species SE50/110. J Biol Chem 277:22853–22862
Acknowledgements
The authors are thankful to the director, CSIR-National Chemical Laboratory, Pune for providing infrastructure facilities. The author acknowledges Council of Scientific and Industrial Research (CSIR), New Delhi for fellowship grant in the form of SRF.
Author information
Authors and Affiliations
Contributions
The author SGD and MSD designed the study and the author RBM performed the experiments and data collection and analysis and manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Human and animal rights
No human and animal experiments were performed in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mawlankar, R.B., Dharne, M.S. & Dastager, S.G. Isolation of potent alpha-glucosidase inhibitor from a novel marine bacterium Arthrobacter enclensis. SN Appl. Sci. 2, 474 (2020). https://doi.org/10.1007/s42452-020-2285-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2285-3