Abstract
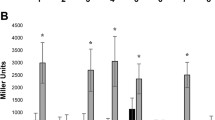
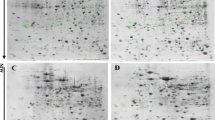
Cupriavidus sp. UYMMa02A is a beta-rhizobia strain of the Cupriavidus genus, isolated from nodules of Mimosa magentea in Uruguay. This strain can form effective nodules with several Mimosa species, including its original host. Genome analyses indicate that Cupriavidus sp. UYMMa02A has a highly conserved 35 kb symbiotic island containing nod, nif, and fix operons, suggesting conserved mechanisms for the symbiotic interaction with plant hosts. However, while Cupriavidus sp. UYMMa02A produces functional nodules and promotes Mimosa pudica growth under nitrogen-limiting conditions, nod genes are not induced by luteolin or exposure to Mimosa spp. root exudate. To explore alternative mechanisms implicated in the Cupriavidus-Mimosa interaction, we assessed the proteomic profiles of Cupriavidus sp. UYMMa02A grown in the presence of pure flavonoids and co-culture with M. pudica plants. This approach allowed us to identify 24 differentially expressed proteins potentially involved in bacterial-plant interaction. In light of the obtained results, a possible model for nod-alternative symbiotic interaction is proposed.










Similar content being viewed by others
Data availability
Data is available upon request to the authors.
References
Amadou C, Pascal G, Mangenot S (2008) Genome sequence of the β-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res 18:1472–1483
Andam CP, Mondo SJ, Parker MA (2007) Monophyly of nodA and nifH genes across Texan and Costa Rican populations of Cupriavidus nodule symbionts. Appl Environ Microbiol 73:4686–4690
Andrews M, Andrews ME (2017) Specificity in legume-rhizobia symbioses. Int J Mol Sci 18:705
Attwood PV, Wieland T (2015) Nucleoside diphosphate kinase as protein histidine kinase. Naunyn Schmiedebergs Arch Pharmacol 388:153–160
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75
Bellés-Sancho P, Liu Y, Heiniger B, von Salis E, Eberl L, Ahrens CH et al (2022) A novel function of the key nitrogen-fixation activator NifA in beta-rhizobia: repression of bacterial auxin synthesis during symbiosis. Front Plant Sci. https://doi.org/10.3389/fpls.2022.991548
Berg P, Joklik WK (1953) Transphosphorylation between nucleoside polyphosphates. Nature 172:1008–1009
Bontemps C, Elliott GN, Simon MF, Dos Reis Júnior FB, Gross E, Lawton RC et al (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Capela D, Carrere S, Batut J (2005) Transcriptome-based identification of the Sinorhizobium meliloti NodD1 regulon. Appl Environ Microbiol 71:4910–4913
Caporali S, De Stefano A, Calabrese C, Giovannelli A, Pieri M, Savini I et al (2022) Anti-inflammatory and active biological properties of the plant-derived bioactive compounds luteolin and luteolin 7-glucoside. Nutrients 14:1–19
Chen CC, Chou TL, Lee CY (2000) Cloning, expression and characterization of L-aspartate β-decarboxylase gene from Alcaligenes faecalis CCRC 11585. J Ind Microbiol Biotechnol 25:132–140
Chen W, Laevens S, Lee T, Coenye T, Vos PD, Mergeay M, Vandamme P (2001) Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735
Chen W-M, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C (2003) Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J Bacteriol 185:7266–7272
Chen W-M, Prell J, James EK, Sheu D-S, Sheu S-Y (2012) Biosynthesis of branched-chain amino acids is essential for effective symbioses between betarhizobia and Mimosa pudica. Microbiology 158:1758–1766
Cheng C-S, Su C-W, Chang W-C, Huang K-C (2014) CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 9:99368
Cunnac S, Boucher C, Genin S (2004) Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J Bacteriol 186:2309–2318
Dall’Agnol RF, Bournaud C, de Faria SM, Bena G, Moulin L, Hungria M (2017) Genetic diversity of symbiotic Paraburkholderia species isolated from nodules of Mimosa pudica (L.) and Phaseolus vulgaris (L.) grown in soils of the Brazilian Atlantic Forest (Mata Atlantica). FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fix027
de Campos SB, Lardi M, Gandolfi A, Eberl L, Pessi G (2017) Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front Microbiol 8:2473
De Meyer SE, Fabiano E, Tian R, Van Berkum P, Seshadri R, Reddy T et al (2015a) High-quality permanent draft genome sequence of the Parapiptadenia rigida-nodulating Cupriavidus sp. strain UYPR2.512. Stand Genom Sci 10:13
De Meyer SE, Parker M, Van Berkum P, Tian R, Seshadri R, Reddy TBK et al (2015b) High-quality permanent draft genome sequence of the Mimosa asperata-nodulating Cupriavidus sp. strain AMP6. Stand Genomic Sci 10:9–11
De Meyer SE, Briscoe L, Martínez-Hidalgo P, Agapakis CM, de-Los Santos PE, Seshadri R et al (2016) Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol Plant Microbe Interact 29:609–619
diCenzo GC, Zamani M, Checcucci A, Fondi M, Griffitts JS, Finan TM, Mengoni A (2019) Multidisciplinary approaches for studying rhizobium–legume symbioses. Can J Microbiol 65:1–33
Dimou M, Venieraki A, Katinakis P (2017) Microbial cyclophilins: specialized functions in virulence and beyond. World J Microbiol Biotechnol 33:1–8
dos Reis FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE et al (2010) Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946
Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155:3166–3175
Estrada-delossantos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp ET, Briscoe L et al (2018) Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (basel). 9:389
Fagorzi C, Bacci G, Huang R, Cangioli L, Checcucci A, Fini M et al (2021) Nonadditive transcriptomic signatures of genotype-by-genotype interactions during the initiation of plant-rhizobium symbiosis. mSystems. https://doi.org/10.1128/mSystems.00974-20
Gambino M, Cappitelli F (2016) Mini-review: biofilm responses to oxidative stress. Biofouling 32:167–178
Garau G, Yates RJ, Deiana P, Howieson JG (2009) Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 41:125–134
Gehlot HS, Tak N, Kaushik M, Mitra S, Chen W-M, Poweleit N et al (2013) An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot 112:179–196
Gil M, Lima A, Rivera B, Rossello J, Urdániz E, Cascioferro A et al (2019) New substrates and interactors of the mycobacterial serine/threonine protein kinase PknG identified by a tailored interactomic approach. J Proteom 192:321–333
Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre J-C et al (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312
Gourion B, Berrabah F, Ratet P, Stacey G (2015) Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci 20:186–194
Guha S, Molla F, Sarkar M, Ibañez F, Fabra A, DasGupta M (2022) Nod factor-independent “crack-entry” symbiosis in dalbergoid legume Arachis hypogaea. Environ Microbiol 24:2732–2746
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580. https://doi.org/10.1016/s0022-2836(83)80284-8
Howieson JG, Robson AD, Ewing MA (1993) External phosphate and calcium concentrations, and Ph, but not the products of rhizobial nodulation genes, affect the attachment of rhizobium meliloti to roots of annual medics. Soil Biol Biochem 25:567–573
Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ (2013) Novel Burkholderia bacteria isolated from Lebeckia ambigua—a perennial suffrutescent legume of the fynbos. Soil Biol Biochem 60:55–64
Huergo LF, Dixon R (2015) The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol Mol Biol Rev 79:419–435
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC et al (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:286–293
Hungria M, Joseph CM, Phillips DA (1991) Rhizobium nod gene inducers exuded naturally from roots of common bean (Phaseolus vulgaris L.). Plant Physiol 97:759–764
Iriarte A, Platero R, Romero V, Fabiano E, Sotelo-Silveira JR (2016) Draft genome sequence of Cupriavidus UYMMa02A, a novel beta-rhizobium species. Genome Announc 4:e01258-e1316
Janczarek M, Rachwał K, Cieśla J, Ginalska G, Bieganowski A (2015a) Production of exopolysaccharide by Rhizobium leguminosarum bv. trifolii and its role in bacterial attachment and surface properties. Plant Soil 388:211–227
Janczarek M, Rachwał K, Marzec A, Grządziel J, Palusińska-Szysz M (2015b) Signal molecules and cell-surface components involved in early stages of the legume–rhizobium interactions. Appl Soil Ecol 85:94–113
Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A et al (2010) The complete genome sequence of cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 5:e10433
Jiménez-Guerrero I, Acosta-Jurado S, del Cerro P, Navarro-Gómez P, López-Baena F, Ollero F et al (2017) Transcriptomic studies of the effect of nod gene-inducing molecules in rhizobia: different weapons, one purpose. Genes (basel). https://doi.org/10.3390/genes9010001
Kakimoto T, Kato J, Shibatani T, Nishimura N, Chibata I (1969) Crystalline l-aspartate β-decarboxylase of Pseudomonas dacunhae. J Biol Chem 244:353–358
Kessler B, de Lorenzo V, Timmis KN (1992) A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet MGG 233:293–301
Klonowska A, Chaintreuil C, Tisseyre P, Miché L, Melkonian R, Ducousso M et al (2012) Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol 81:618–635
Klonowska A, Melkonian R, Miché L, Tisseyre P, Moulin L (2018) Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic ev. BMC Genom 19:105
Klonowska A, Moulin L, Ardley JK, Braun F, Gollagher MM, Zandberg JD et al (2020) Novel heavy metal resistance gene clusters are present in the genome of Cupriavidus neocaledonicus STM 6070, a new species of Mimosa pudica microsymbiont isolated from heavy-metal-rich mining site soil. BMC Genom 21:214
Kornberg HL (1966) The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Laemmli U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lardi M, Liu Y, Purtschert G, de Campos SB, Pessi G (2017) Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes (basel). 8:389
Lascu I, Gonin P (2000) The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr 32:237–246
Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, Boatwright JS et al (2015) Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol Ecol 91:1–17
Lindström K, Mousavi SA (2020) Effectiveness of nitrogen fixation in rhizobia. Microb Biotechnol 13:1314–1335
Lipa P, Vinardell JM, Kopcińska J, Zdybicka-Barabas A, Janczarek M (2018) Mutation in the pssZ gene negatively impacts exopolysaccharide synthesis, surface properties, and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Genes (basel). 9:369
Liu X, Wei S, Wang F, James EK, Guo X, Zagar C et al (2012) Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol 80:417–426
Liu Y, Bellich B, Hug S, Eberl L, Cescutti P, Pessi G (2020) the exopolysaccharide cepacian plays a role in the establishment of the Paraburkholderia phymatum—Phaseolus vulgaris symbiosis. Front Microbiol 11:1600
Lu Q, Park H, Egger LA, Inouye M (1996) Nucleoside-diphosphate kinase-mediated signal transduction via histidyl-aspartyl phosphorelay systems in Escherichia coli. J Biol Chem 271:32886–32893
Mailloux RJ, Bériault R, Lemire J, Singh R, Chénier DR, Hamel RD, Appanna VD (2007) The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2:e690
Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C et al (2010) Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol 8:e1000280
Marchetti M, Catrice O, Batut J, Masson-Boivin C (2011) Cupriavidus taiwanensis bacteroids in Mimosa pudica Indeterminate nodules are not terminally differentiated. Appl Environ Microbiol 77:2161–2164
Martinez A, Kolter R (1997) Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179:5188–5194
Masson-Boivin C, Giraud E, Perret X, Batut J (2009) Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17:458–466
Meleady P (2018) Two-dimensional gel electrophoresis and 2D-DIGE. Methods Mol Biol 1664:3–14
Miché L, Moulin L, Chaintreuil C, Contreras-Jimenez JL, Munive-Hernández JA, del Carmen Villegas-Hernandez M et al (2010) Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that nod-independent stem nodulation is not restricted to photosynthetic bradyrhizobia. Environ Microbiol 12:2152–2164
Miller JH (1972) Assay of B-galactosidase In: Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Moskowitz GJ, Merrick JM (1969) Metabolism of poly-β-hydroxybutyrate. II. Enzymic synthesis of D-(-)-β-hydroxybutyryl coenzyme A by an enoyl hydrase from Rhodospirillum rubrum. Biochemistry 8:2748–2755
Moulin L, Klonowska A, Caroline B, Booth K, Vriezen JAC, Melkonian R et al (2014) Complete genome sequence of Burkholderia phymatum STM815(T), a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand Genom Sci 9:763–774
Mozejko-Ciesielska J, Mostek A (2019) A 2D-DIGE-based proteomic analysis brings new insights into cellular responses of Pseudomonas putida KT2440 during polyhydroxyalkanoates synthesis. Microb Cell Fact 18:1–13
Nies DH (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339
Nyström T, Neidhardt FC (1994) Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol 11:537–544
Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144
Orban K, Finkel SE (2022) Dps is a universally conserved dual-action DNA-binding and ferritin protein. J Bacteriol 204:1–23
Parker MA, Wurtz AK, Paynter Q (2007) Nodule symbiosis of invasive Mimosa pigra in Australia and in ancestral habitats: a comparative analysis. Biol Invasions 9:127–138
Pereira-Gómez M, Ríos C, Zabaleta M, Lagurara P, Galvalisi U, Iccardi P et al (2020) Native legumes of the Farrapos protected area in Uruguay establish selective associations with rhizobia in their natural habitat. Soil Biol Biochem 148:107854
Platero R, James EK, Rios C, Iriarte A, Sandes L, Zabaleta M et al (2016) Novel Cupriavidus strains isolated from root nodules of native Uruguayan Mimosa species. Appl Environ Microbiol 82:3150–3164
Prell J, Poole P (2006) Metabolic changes of rhizobia in legume nodules. Trends Microbiol 14:161–168
Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, Poole PS (2009) Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc Natl Acad Sci USA 106:12477–12482
Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS (2011) Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12:R106
Rodríguez-Esperón MC, Eastman G, Sandes L, Garabato F, Eastman I, Iriarte A et al (2022) Genomics and transcriptomics insights into luteolin effects on the beta-rhizobial strain Cupriavidus necator UYPR2.512. Environ Microbiol 24:240–264
Saad MM, Crèvecoeur M, Masson-Boivin C, Perret X (2012) The type 3 protein secretion system of Cupriavidus taiwanensis strain LMG19424 compromises symbiosis with Leucaena leucocephala. Appl Environ Microbiol 78:7476–7479
Salehi B, Venditti A, Sharifi-Rad M, Kręgiel D, Sharifi-Rad J, Durazzo A et al (2019) The therapeutic potential of Apigenin. Int J Mol Sci 20:1305
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Santos MR, Marques AT, Becker JD, Moreira LM (2014) The Sinorhizobium meliloti EmrR regulator is required for efficient colonization of Medicago sativa root nodules. Mol Plant Microbe Interact 27:388–399
Schlaman HRM, Okker RJH, Lugtenberg BJJ (1992) Regulation of nodulation gene expression by nodD in rhizobia. J Bacteriol 174:5177–5182
Schmidt PE, Broughton WJ, Werner D (1994) Nod factors of Bradyrhizobium-japonicum and Rhizobium sp. NGR234 induce flavonoid accumulation in soybean root exudate. Mol Plant Microbe Interact 7:384–390
Shankar S, Kamath S, Chakrabarty AM (1996) Two forms of the nucleoside diphosphate kinase of Pseudomonas aeruginosa 8830: altered specificity of nucleoside triphosphate synthesis by the cell membrane-associated form of the truncated enzyme. J Bacteriol 178:1777–1781
Skagia A, Zografou C, Vezyri E, Venieraki A, Katinakis P, Dimou M (2016) Cyclophilin PpiB is involved in motility and biofilm formation via its functional association with certain proteins. Genes Cells 21:833–851
Stancik IA, Šestak MS, Ji B, Axelson-Fisk M, Franjevic D, Jers C et al (2018) Serine/threonine protein kinases from bacteria, archaea and Eukarya share a common evolutionary origin deeply rooted in the tree of life. J Mol Biol 430:27–32
Suzuki S, Aono T, Lee KB, Suzuki T, Liu CT, Miwa H et al (2007) Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl Environ Microbiol 73:6650–6659
Taulé C, Zabaleta M, Mareque C, Platero R, Sanjurjo L, Sicardi M et al (2012) New betaproteobacterial Rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl Environ Microbiol 78:1692–1700
Thomloudi E-E, Skagia A, Venieraki A, Katinakis P, Dimou M (2017) Functional analysis of the two cyclophilin isoforms of Sinorhizobium meliloti. World J Microbiol Biotechnol 33:28
Wu PS, Yen JH, Kou MC, Wu MJ (2015) Luteolin and apigenin attenuate 4-hydroxy-2-nonenal-mediated cell death through modulation of UPR, Nrf2-ARE and MAPK pathways in PC12 cells. PLoS ONE 10:1–23
Yu H, Rao X, Zhang K (2017) Nucleoside diphosphate kinase (Ndk): a pleiotropic effector manipulating bacterial virulence and adaptive responses. Microbiol Res 205:125–134
Zheng J, Wang R, Liu R, Chen J, Wei Q, Wu X et al (2017) The structure and evolution of beta-rhizobial symbiotic genes deduced from their complete genomes. Immunome Res 13:131
Acknowledgement
The authors would like to thank to Prof. Catherine Masson-Boivin for providing plasmids pCZ388 and pCBM01.
Funding
Funding was provided by Agencia Nacional de Investigación e Innovación (grant no. FCE_1_2014_1_104338, FCE_1_2017_1_136082, FCE_1_2019_1_156520), Programa de desarrollo de las ciencias básicas, PEDECIBA (grant no. 2018), and FONTAGRO (grant no. ID 30).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rodríguez-Esperón, C., Sandes, L., Eastman, I. et al. Nodulation in the absence of nod genes induction: alternative mechanisms involved in the symbiotic interaction between Cupriavidus sp. UYMMa02A and Mimosa pudica. Environmental Sustainability 6, 383–401 (2023). https://doi.org/10.1007/s42398-023-00286-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42398-023-00286-5