Abstract
Nowadays, metal oxide nanoparticles (MO NPs) are powerful tools for biological applications due to their distinctive features. Moreover, the biological efficacy of multimetallic NPs is more fascinating because of their structural modifications and synergistic effects. This study utilized the one-pot green route to fabricate trimetallic ZnO-MgO-CuO (ZMC) NPs employing a greener reducing agent from Artemisia abyssinica leaf extract (AALE). The crystal structure, size, compositions, shapes, and external topology of ZMC NPs were characterized by Fourier transform infrared (FTIR), UV–Visible (UV–vis), X-ray diffraction (XRD), scanning electron microscopy (SEM), energy-dispersive X-ray (EDX), and transmission electron microscopy combined with selected area electron diffraction (TEM/HRTEM-SAED). The outcomes suggested that the bio-prepared ZMC NPs are highly crystalline and have hexagonal structures lattice with monoclinic symmetry and spherical morphology with average crystalline and particle sizes of 14.67 and 15.13 nm, respectively. Using MTT assay, the bio-prepared ZMC NPs demonstrated high inhibition percentage (94.37 ± 0.14 at 250 mg/mL) with an IC50 value of 24.83 mg/mL for MCF-7 cell lines. The in-vitro antibacterial potential of ZMC NPs has been evaluated against four bacterial (Gram-positive and Gram-negative) strains and has demonstrated the highest inhibition zone (35 ± 0.03 mm) against the S. aureus strain and the lowest inhibition zone (31 ± 0.11) against the E. coli strain. Moreover, ZMC NPs have also shown strong molecular binding interactions with amino acids of estrogen receptor (ERα), S. aureus, and E. coli with binding energies of − 9.85, − 12.31, and − 6.04 kcal/mole, respectively.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The multi-applications of metal oxide nanoparticles (MO NPs) in various fields including catalysis, energy, environment, agriculture, medicine and biological sciences, etc. gained consideration from scientific societies recently [1]. The size-dependent properties of MO NPs such as high surface area, enhanced solubility, and reactivity as well as optical, electronic, and magnetic properties, hold a pledge to their applications in biological sciences [2]. Moreover, the capacity of MO NPs to generate a high number of ROS (reactive oxygen species), with great selectivity and lower sensitivity to normal cells, urges their use in nanobiotechnology [3].
Among many biologically benign MO NPs that have been introduced into clinical fields, ZnO, and CuO nanoparticles have disclosed several benefits such as bioavailability, biocompatibility, and cost-effectiveness in biological applications [4,5,6]. Due to the semi-conductive nature and surface crystal defects of these NPs the electrons freely move from the valance band to conduction bands and hence could react with oxygen species to form free radicals which cause oxidative damage of in bacterial and cancerous cells. ZnO NPs is more appropriate in cancer therapy due to its inherent electrostatic nature and selective toxicity [7, 8]. ZnO nanoparticles have exhibited significant anticancer efficacies against a variety of cancer types, including lung cancer [9], breast cancer [10], ovarian cancer [11], and colorectal cancer [12]. The anticancer potency of ZnO NPs is mainly through zinc-mediated protein activity disequilibrium and oxidative stress [13, 14]. CuO NPs have shown enhanced anticancer efficacy against breast cancer [15], hepatocellular carcinoma [16], lung cancer [17], and cervical cancer [18]. The anticancer mechanism of CuO NPs is mainly through genotoxicity and apoptotic death in cancerous cells due to its ability to generate enhanced ROS [19]. The exceptional features of MgO NPs such as biocompatibility, recycling activity, low density, hygroscopic nature, and good functionality make it a promising anticancer remedy [20,21,22]. Moreover, nano-metal oxides of zinc, copper and magnesium have been broadly utilized in microbial infections [23,24,25]. However, the integration of ZnO, MgO, and CuO NPs together is preferred over monometallic counterparts due to their immensely improved synergistic effects including enriched surface area, multiple reactive sites, higher charge flow, and mass transfer for biological application [26,27,28].
For the synthesis of trimetallic oxide nanoparticles (TMO NPs), chemical, physical, and biological routes have all been used [29,30,31,32]. The fabrication of TMO NPs from biological entities has many benefits over other techniques. The bioreduction approach is effective, economical, and environmentally benign prohibiting the assembly of undesirable or toxic chemicals [33,34,35]. The green fabrication of MMO NPs has been implemented to accommodate diverse bio-systems, including algae, fungi, bacteria, and plant extracts. Moreover, using plant extracts has a low cost of handling, safety, and simple protocol to create TMO NPs at a large scale relative to bacteria and/or fungi-assisted fabrication [36,37,38,39]. Various secondary metabolites from the plant extracts, including phenolic compounds, flavonoids, alkaloids, terpenoids, etc., contain functional groups such as NH2, SH, COOH, C=O, and OH, contribute to the strong reduction and stabilization of NPs [40]. Moreover, medicinal plant mediated synthesized NPs could also exhibit enhanced biological activity due to the synergistic effects of phytochemicals [25, 41].
Cancer and infectious diseases have become a global burden that causes international health, economic, and communal crises [42, 43]. Due to their unpredictable spread, infectious diseases have deleterious impacts worldwide, including high mortality rates, enormous burdens, and impairment [43, 44]. However, several anticancer drugs have been reported recently, majority of them revealed different encounters in clinical use, like poor water solubility, limited membrane transport capacity, and swift clearance and degradation problems in the bloodstream during clinical use [45]. The path of physiological response to counter them as well as the cytotoxicity to a healthy body of conventional chemotherapy recently became the main focus of researchers [46]. Antibiotics are one of human medicine's most extensively utilized treatment techniques to fight pathogenic microorganisms [47]. However, changes in the ability of microorganisms to resist antibiotics, either by inactivating them or limiting their therapeutic efficacy, lead to microbial resistance, also known as multi-drug resistance [48].
In this study, Artemisia abyssinica leaf extract (AALE) was used as a greener reducing and stabilizing agent for the synthesis of ZMC NPs. Artemisia abyssinica belongs to the family Asteraceae indigenous medicinal plant regularly used for the treatment of different diseases traditional as well as modern medicine of Ethiopia [49]. AA was known to treat inflammation, cold, amenorrhea, headache, colic, anorexia, fever, and dysmenorrhea traditionally [50]. Moreover, the plant has also been reported as an antimalarial, antiparasitic, antirheumatic, anti-inflammation, antitumor, and antioxidant agent [51,52,53]. Essential oils, tannins, terpenoids, saponins, polyphenols, alkaloids, and flavonoids are the most common phytochemicals reported from the AALE [54, 55].
Therefore, in this study we hereby report a novel, eco-benign synthesis of trimetallic ZMC NPs using biomolecules from AALE for the first time. For the characterizations of biosynthesized ZMC NPs the advanced techniques including UV–visible, XRD, FTIR, SEM, EDX, TEM/HRTEM and SAED were used. Furthermore, comprehensive studies of in-vitro cytotoxicity and antibacterial potentials as well as in-silico molecular docking analysis of biosynthesized ZMC NPs were also presented.
2 Materials and Methods
2.1 Chemicals and Media
Magnesium nitrate hexahydrate (Mg(NO3)2·6H2O, 99.8%), zinc nitrate tetrahydrate (Zn(NO3)2·4H2O, 99.9%), copper nitrate trihydrate, (Cu(NO3)2·3H2O, 99.8%), ethanol (C2H6O, purity ≥ 99.9%), and sodium hydroxide (NaOH, 97%), dimethyl sulfoxide (DMSO), ascorbic acid and Chloramphenicol, Sigma Aldrich, were purchased from Addis Ababa, Ethiopia. Mueller Hinton broth (MHB), Sabouraud Dextrose Agar (SDA), Potato Dextrose Agar (PDA), 2,2-diphenyl-1-picryl-hydrazyl-hydrate (DPPH), 3-(4,5-dimethylthiazol-2-yl)-2, were procured from institute of public health (IPH), Addis Ababa, Ethiopia. Dulbecco's Modified Eagle Medium (DMEM), Fetal Bovine Serum (FBS), 3-(4,5-dimethylthiazol-2-yl)-2, 5diphenyltetrazolium bromide (MTT), PenStrep, Trypsin, Spectramax I3X, CO2 incubator, and Doxorubicin (Invitrogen, USA) where obtained from, India.
2.2 Bacteria and Cancer Cell Lines
Bacteria strains, Escherichia coli (E. coli, ATCC 25922), Pseudomonas aeruginosa (P. aeruginosa, ATCC 27823), Streptococcus pneumoniae (S. pneumoniae, ATCC 11778), and Staphylococcus aureus (S. aureus ATCC 25923), were obtained from the Institute of Public Health, Addis Ababa, Ethiopia. Breast cancer (MCF-7) cells and peripheral blood mononuclear cells (PBMCs) (Invitrogen, USA) were acquired from National Center for Cell Sciences (NCCS), India.
2.3 Preparation of Artemisia abyssinica Leaf Extract (AALE)
The preparation of AALE was done according to our previous work with certain modifications [54]. The leaves part of the medicinal plant Artemisia abyssinica was collected from Tiyo Woreda, Arsi Zone, Tiyo Woreda, 7°45′55'' and 8°02′02''N latitude and 38°56′42'' to 39°18′31''E longitude, Oromia Region, Ethiopia, which is 175 km Southeast of Addis Ababa with an elevation ranges of 1850–4050 m [56]. The obtained leaves were washed with tap and distilled water (dH2O) repeatedly and dried for 15 days in the dark to remove moisture content. After grinding with a mechanical grinder, 10 g of powdered leaves were added with 100 mL of 50% ethanol (water and ethanol, 1:1 v/v) containing 250 mL conical flasks, and then the mixtures were shaken for 1 and 1/2 h at 120 rpm and 25 °C in a mechanical shaker and then heated for 50 min at 60 °C on a magnetic stirrer. After centrifuging the drift to produce a clear solution, it was chilled at room temperature (RT) overnight before the resulting solutions were filtered through Whatman filter paper. Finally, a clear brown color extract was obtained and stored at 4 °C for further study.
2.4 Bio-Preparation of ZMC NPs
Bio-preparation of ZMC NPs was carried out by mixing 10 mM of each salts [Cu(NO3)2·3H2O, Mg(NO3)2·6H2O, and Zn(NO3)2.4H2O] solution in a 1:1:1 ratio with AALE in a proportion of 3:1 (v/v). According to optimizations of our previous study [54], the solution was adjusted at pH 5, and heated for 1 h at 70 °C on a hot plate containing a magnetic stirrer. The light blue to deep brown color change has been observed for the spontaneous reduction and formation of MZC NPs. The reaction mixture was sonicated for 1 h at RT at 60 rpm to retain the particle dispersion. The resultant solution was then centrifuged at 6000 rpm for 30 min. The pellets were repeatedly cleaned with distilled water (dH2O) following ethanol to remove the impurities and then oven-dried for 12 h at 80 °C, annealed for 2 h at 400 °C; the fine deep brown powder was obtained and stored in protective sample holders at 4 °C for further analysis [57].
2.5 Characterizations
For the characterization of the biosynthesized ZMC NPs various techniques such as UV–visible (UV–vis) spectroscopy, Fourier transform infrared (FTIR) spectroscopy, X-ray diffraction (XRD), scanning electron microscopy (SEM), energy dispersive X-ray analysis (EDAX), transmission electron microscopy (TEM), and selected area electron diffraction (SAED) were used.
2.5.1 Absorption Edge Determination
For the estimation of absorption edge of the as biosynthesized ZMC NPs, UV–vis diffuse absorption was measured using SP65 spectrophotometer (Shimadzu UV–vis, SM-1600) at a scanning range of 200–800 nm.
2.5.2 FTIR Analysis
Fourier transform-infrared spectroscopy (Perkin Elmer FT-IR, Spectrum 65) using KBr pellets in the range of 400–4000 cm−1 was used to study the responsible functional groups for the reduction and stability of biosynthesized ZMC NPs and their chemical bonding behavior.
2.5.3 XRD Study
The X-ray diffraction (XRD, Shimadzu, and XRD-7000) equipped with a Cu target for generating a Cu Kα radiation (wavelength 1.5406 Å) was used to study the actuality and crystallinity of as synthesized ZMC NPs. The measurements were made at room temperature and the accelerating voltage and the applied current were 40 kV and 30 mA, respectively. The instrument was operated under step scan type with step time and degree (2θ) of 1 s and 0.020°, respectively over 2θ range of 10° to 80°.
2.5.4 SEM/EDX Analysis
The morphology and composition of the biosynthesized ZMC NPs were characterized by scanning electron microscopy (SEM, JEOL-JSM 6500F, made in Japan); SEM coupled energy-dispersive X-ray spectroscopy (EDX).
2.5.5 TEM/HRTEM/SAED Analysis
The internal morphology, particle size, and interplanner distance of ZMC NPs were characterized by transmission electron microscope (TEM/HR-TEM/SAED, Tecnai F20 G2, Philips, and the Netherlands).
2.6 Cytotoxicity Test
The standard method was used to culture the MCF-7 and PBMC cells [58]. The cells were given injections of 10% (v/v) FBS, streptomycin (100 g/mL), and penicillin (100 IU/mL) until congestion was achieved while being cultured in DMEM with 5% CO2 at 37 °C. Following that the matured cells were isolated with cell separating solution (0.2% trypsin, 0.02% EDTA, 0.05% glucose in PBS (Fetal Bovine Serum), then centrifuged and tested for viability. Furthermore, 5 × 104 cells /well were cultivated in a 96-well plate and nurtured for 24 h at 37 °C, under a 5% CO2 incubator.
The cytotoxicity of ZMC NPs and standard drug (Doxorubicin) against MCF-7 and PBMC cells were evaluated using MTT assay [59, 60]. The standard cells were trypsinized and adjusted with 5 × 104 cells/ml to count cells using respective media containing 10% FBS (v/v). The diluted cell suspension (5 × 104 cells/well) was poured into the 96-wells of the microliter plate in a quantity of 100 µL. The test solutions in the wells were rejected after incubation and 0.05 mg MTT was poured into the wells. The plates were kept for 4 h at 37 °C under a 5% CO2 atmosphere. The supernatants were removed from each plate and shaken gently and then 100 mL of DMSO was added to dissolve the formed formazan. The absorbance was recorded with a microplate reader at 570 nm. The growth inhibition percent (%) of the cells was computed using Eq. (1) and IC50 values were determined by using dose–response curves.
where, OD stands optical density.
2.7 Antibacterial Activity Test
The antibacterial efficacy of the bio-prepared ZMC NPs was evaluated using the agar disc diffusion method [61]. Mulberry Hinton Broth and Muller Hinton Agar were used as liquid and solid mediums, respectively. Using 0.5 McFarland standard the density of bacteria was adjusted and wiped on a plate covered with Muller Hinton agar from a suspension of fresh bacterial cultures such as E. coli, S. pneumoniae, S. aureus, and P. aeruginosa mixed to the liquid medium to obtain turbidity. The disks were soaked with 50 μL of (12.5, 25, 50, 100, and 200 μg/mL) ZMC NPs. Chloramphenicol (positive control) and DMSO (negative control) were used by impregnated standard discs (30 μg) as standard antibiotics. The cultures were incubated for 24 h at 37 °C, and then the zone of inhibition (ZOI) was recorded with a ruler (in mm) to ascertain the bacterial strains liability to the samples. Each trial was carried out in triplicate and reported in mean standard deviation using statistical analysis software (SPSS) (version 20).
2.8 Molecular Docking Study
The molecular interaction of ZMC NPs and the standard drugs (doxorubicin and chloramphenicol) were evaluated against the binding sites of estrogen receptor alpha (ERα; PDB: 5GS4), dihydrofolate reductase (PDB: 2w9h) (DHR), and DNA gyrase B (PDB: 6F86) using AutoDock 4.2.6 (MGL tools 1.5.7) programs. Estrogen receptor alpha, dihydrofolate reductase, and DNA gyrase B are the important protein targets of MCF-7, S. aureus, and E. coli respectively [62, 63]. The geometry of ZMC NPs and standard drugs were augmented using the B3LYP-GD3/6–311 + + G(d,p)/LanL2DZ approach before the molecular docking investigation, and then translated to PDB files applying GaussView. The protein database was used to download the crystal structures of the targeted proteins. The structures of targeted proteins were then processed by adding polar hydrogen and cofactors, as well as eliminating the co-crystallized ligand and water molecules according to the AutoDock 4.2.6 (MGL tools 1.5.7) program. After cleaning the protein, only polar hydrogen and the Kollman charges were introduced. Nonpolar hydrogen atoms were merged and Gasteiger partial atomic charges were assigned to the compounds. The grid center coordinates were 65, 65, and 65 pointing in the respective x, y, and z directions with a grid point spacing of 0.375 Å. The grid box was centered at – 12.055, – 10.491, and 5.964 Å. Using 100 alternative conformations each standard drug and ZMC NPs were evaluated. Discovery Studio Visualizer was employed to visualize the binding interactions between ZMC NPs and standard drugs with the target receptors exhibiting the lowest binding free energies.
2.9 Statistical Data Analysis
The experimental results were computed using the one-way analysis of variance (ANOVA) function of the statistical package for social science (SPSS) version 20 and presented as mean ± standard deviation for triplicate experiments. ImageJ (imajej153-win java8 imagej.exe), Gatan Microscopy Suite software (GMS 64bit) version 2.x, and Origin software (Originpro 9.0 64bit) were also utilized for data processing.
3 Results and Discussion
3.1 One-Pot Bio-Preparation of ZMC NPs
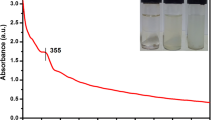
Trimetallic ZMC NPs were synthesized by one-pot green preparation method using ethanollic (1:1, v/v) AALE. The addition of AALE to the mixed salts solution after 1 h of reaction at 70 °C on a hot plate resulted in color change from reddish-brown to a black-brown. Initially, the formation of ZMC NPs was confirmed qualitatively by the steady color change from reddish-brown to a black-brown (Fig. 1). Furthermore, the UV–visible spectroscopy was used to test the formation of ZMC NPs as presented in Fig. 2. The significant surface Plasmon resonance (SPR) peak at 367 nm confirmed the bioreduction of salts [Cu(NO3)2·3H2O, Mg(NO3)2·6H2O, and Zn(NO3)2·4H2O] solution to ZnO–MgO–CuO (ZMC) NPs [16, 64].
Figure 1 illustrates the mechanism of interactions between precursor salts and phytochemicals from the extract of A. abyssinica during the synthesis of MZC NPs, whereas Fig. 2 presents the UV-visible spectra. The main secondary metabolites in A. abyssinica extract are phenolic compounds such as polyphenols, alkaloids, and flavonoids used as reducing and stabilizing ligands [54]. Polyphenols are potential agents that could participate as reducing and capping ligands among the phytochemicals mentioned, that are also illustrated in the FTIR study (Fig. 3). However, polyphenols most commonly contain phenolic acids, stilbenes, and lignans [65]. Among the phenolic acids accounts for the majority of polyphenols in most reported works of literature and also are those with the most active sites than the others [66]. In light of this, a proposed reaction scheme for forming ZMC NPs is presented as follows.
3.2 UV–visible Study
UV–visible (UV–vis) spectroscopy is an essential technique used to determine NPs physico-chemical properties, electronic structure, and optical activities. 5 mM of AALE and ZMC NPs aqueous solutions were prepared and their respective absorption peaks were measured using UV–visible spectroscopy. AALE revealed various absorption peaks at 254, 360, and 402 nm. This could be due to various active biomolecules present in AALE, which are responsible for the reduction and stability of synthesized nanoparticles [67]. The biosynthesized ZMC NPs revealed the SPR band at 367 nm. The interaction between the light wave and the ZMC NPs free electrons may be what causes the SPR absorption [68, 69]. According to recent research, spherical NPs only display one SPR band, whereas anisotropic particles evince two or more SPR bands [70]. Hence the single SPR peak in the UV–vis spectrum of ZMC NPs illustrates the formation of iso-morphological particles [69]. The symmetrical shape of SPR bands also suggests that the biosynthesized ZMC NPs have well-dispersed and uniform sizes [54, 71]. Since non-uniform particles will have a broad absorption peak with a split Plasmon band [72]. Moreover, the higher intensity in the UV–vis spectra of ZMC NPs could also use to carry out the quantitative analysis of yields of nanoparticles [73]. Hence absorbance of the peak has direct relation to the concentration of the absorbing species in the medium. Therefore, the UV–vis spectra of ZMC NPs (Fig. 2) demonstrated the formation of well dispersed, iso-morphological particles with high yield.
3.3 FTIR Study
FTIR spectroscopy is a helpful tool for identifying functional groups of active phytoconstituents that serve as reductants and stabilizers in the bio-fabrication of ZMC NPs. FTIR spectra of the bio-fabricated ZMC NPs and AALE are presented in Fig. 3. The absorption band positions (Table 1) at 3436, 2914, 2072, 1392, 1268, 1076, and 512 cm−1 represent the FTIR spectra of the AALE. The bending and stretching vibrational frequencies of phenolic –OH is presumed to be involved inthe broadband at 3436 cm−1 [74]. The narrow peak at 2914 cm−1 is also ascribed and sharp peak at 2072 cm−1 are due to the bending and stretching frequency of C-H from vinyl groups C≡C from terminal alkyl groups respectively [54, 74]. The prominent peak at 1392 cm−1 could be accountable for the stretching vibration of C=O of polyphenols. The intense peaks at 1268, and 1076 cm−1 were due to stretching vibrations of aromatic C-O and N–H from phenolic groups respectively [75, 76]. The broad peak at 512 cm−1 could be connected to the esters C–O or C–O–C stretching [75].
Bio-prepared ZMC NPs nanoparticles have shown FTIR spectra absorption band positions of 3354–3206, 2914, 2072, 1381, 1105, 833, 614, and 524 cm−1 (Fig. 3 and Table 1). The broad peak at 3354–3206 cm−1 is attributed to the stretching frequency of phenolic O–H from extract, which is used as a reducing and capping ligand. The broad peak at 2914 cm−1 was due to the stretching vibration of methylene from vinyl groups. The demanding peak at 2072 cm−1 was due to the stretching vibration of C≡C from the terminal alkyl group [77]. The intense peak at 1381 was due to the C=O vibrational frequency from phenolic compounds during the production of ZMC NPs. The prominent peaks at 1105, 833, and 614 cm−1 correspond to the stretching vibrations of the Cu–O, Zn–O, and Mg-O bonds [64, 77, 78]. The broad peak at 588 cm−1 could be assigned due to the stretching of C–O–C. In general, FT-IR spectra of synthesized materials revealed that NPs are coated with active phytoconstituents, particularly with the O–H, C=O, and C–N residues of phenolic compounds and alkaloids. O–H, C=O, and C-N residues can bond with metal by coating their surface and minimizing agglomeration, which are crucial for stabilization [75].
3.4 XRD Study
The X-ray diffractometer (XRD) pattern of the bio-fabricated ZMC NPs is illustrated in Fig. 4. The prominent peaks at 27.75°, 31.73°, 34.36°, 36.12°, 38.74°, 42.98°, 47.58°, 57.10°, 61.82°, 73.06° and 78.54° associated with the Miller indices (hkl) values of (110), (100), (002), (002), (111), (200), (102), (110), (220), (311), and (222) respectively. The typical diffraction peaks at (100), (002), (102), and (110) planes with 2θ values 31.73°, 34.36°, 47.58°, and 57.10° are associated with the crystalline structure of zinc oxide NPs with the (ICSD card No. 00–036-1451, Zincite-P63mc) [79]. The characteristic diffraction peaks at (002), (111), (220), and (311) planes with peak positions of 36.12°, 38.74°, 61.82°, and 73.06° are associated with the CuO NPs with the (ICSD card No. No.00–048-1548, Tenorite-C2/c) [80]. The prominent diffraction peaks at (110), (200), and (222) planes with diffraction positions of 18.57°, 42.98°, and 78.54° are also associated with the MgO NPs with the (ICSD card No. 00–045-0946, Periclase-Fm-3 m) [81]. These XRD spectra results revealed the successful synthesis of highly crystalline ZMC NPs. The sharper to broader diffraction peaks are observed, and the displayed (101), (100), (002), (002), (111), (200), (102), (110), (220), (311), and (222) planes are indicating the formation of the hexagonal structures lattice with monoclinic symmetry [82].
The mean crystallite size of the biosynthesized ZMC NPs was computed from the full width at half-maximum (FWHM) of the diffraction peaks at the (101), (100), (002), (200), and (311) planes. The Scherrer’s equation was applied to approximate the crystallite size of the synthesized LDHs from the significant diffraction peaks utilizing the equation. Using the Scherrer’s Eq. (2), the average crystalline size was calculated to be 14.67 nm.
where, ‘D’ is the average crystal size, ‘λ’ is the wavelength of the X-ray radiation (Cu Kα = 0.15418 nm), ‘0.9’ stands the Scherrer’s constant ‘K’, and ‘β’ is FWHM.
3.5 SEM–EDX Analysis
The surface topology, structure, and composition of the bio-fabricated ZMC NPs were revealed using SEM–EDX data. The SEM micrographs of ZMC NPs at different magnifications were presented in Fig. 5a, b. The micrographs indicate that the bio-prepared ZMC NPs were found to be flower-like aggregates with spherical morphology. Furthermore, the particles are dispersed across the surface more uniformly with a low level of agglomeration/clustered forms, and that may be allied with the presence of phytochemicals from the extract of Artemisia abyssinica.
Figure 5c demonstrates the EDX spectrum of bio-prepared ZMC NPs. The result indicates the presence of zinc, copper, magnesium, and oxygen at high percentages and carbon and nitrogen at minimum extents. The weight percentages (%) of the zinc, copper, magnesium, and oxygen were 22.46, 20.56, 13.87, and 26.50%, respectively. The compositions of these elements clearly indicate the successful synthesis of pure ZMC NPs. However, 13.30 and 3.31% of carbon and nitrogen imply some impurity level due to the consisting active biomolecules binding with the surface of ZMC NPs. The intense peaks in (Fig. 6c) around 0.5, 1.2, 1.4, 8.2, and 8.7 keV energies also confirm the crystalline nature of bio-prepared ZMC NPs.
3.6 TEM-HRTEM-SAED Analysis
The morphology, particle size, and crystallinity of bio-fabricated ZMC NPs were further studied by TEM-HRTEM-SAED images (Fig. 6a–f). The bio-prepared ZMC NPs have well-defined spherical morphology with sizes ranging from 4 to 26 nm and an average particle size of 15.13 nm as presented in Fig. 6a, b. The presence of small particles with the size of 4 nm demonstrates the effectiveness of biomolecules from the extract of Artemisia abyssinica as capping and stabilizing agents. The nine major circular concentric circles on the SAED pattern (Fig. 6c) correspond to (110), (100), (002), (002), (111), (200), (102), (110), (220) and (311) crystal planes of ZMC NPs. Colored concentric circles also correlated the nine most noticeable patterns found in the XRD measurements in (Fig. 4). The average inter-planer spacing (IPS) values of 0.276, 0.261, and 0.211 nm (Fig. 6d), which was computed from profile IFFT in (Fig. 6e–g) well associated with the (002), (100) and (110) planes of CuO, ZnO, and MgO NPs respectively. The stacking faults on the HRTEM (IFFT) image and the presence of bright circular spots in the SAED pattern confirms the preparations of polycrystalline ZMC NPs [83].
3.7 Biological Efficacy
The in-vitro cytotoxicity and antibacterial efficacy of bio-prepared ZMC NPs was scrutinized against MCF-7 cell lines and pathogenic bacterial strains, respectively. The in-silico molecular docking efficiency of biosynthesized ZMC NPs also estimated against amino acids of estrogen receptor (ERα), S. aureus, and E. coli, respectively.
3.7.1 Cytotoxicity Test
The in-vitro cytotoxicity of bio-prepared trimetallic ZMC NPs and doxorubicin (the standard drug) have been assessed against PBMCs and MCF-7 cell lines. Evaluating the cytotoxicity on normal human cell lines was thought to be the initial step in determining the safety of the fabricated NPs [84]. The cytotoxicity results of the bio-prepared ZMC NPs and doxorubicin against MCF-7 and PBMC are presented in Table 2 and Figs. 7 and 8. The obtained results corroborated that the % inhibition of ZMC NPs against PBMC cell lines was 55.36 ± 0.11. Doxorubicin has shown a % inhibition value of 61.86 ± 0.02 against PBMC cell lines. ZMC NPs have shown remarkably lower cytotoxicity against PBMC cell lines than the standard drug. The IC50 of ZMC NPs and Doxorubicin were 227.69, and 175.56 mg/mL, respectively. The obtained high value of IC50, which is ≥ 90 μg/mL, confirmed that ZMC NPs are non-cytotoxic to the PBMC cell lines [85]. This may be due to the synthesis of NPs from biological precursors and more physiologically functioning (magnesium, zinc, and copper) metal ions, making them biocompatible and less cytotoxic to normal cells.
The bio-prepared ZMC NPs and doxorubicin were also evaluated against MCF-7 cell lines. The results in Table 2 and Figs. 7 and 8 have shown a decline in cell viability in a concentration-dependent manner. The highest inhibition percent of ZMC NPs and doxorubicin were 94.37 ± 0.14 and 86.62 ± 0.04 at 250 mg/mL, respectively. ZMC NPs and doxorubicin have also revealed the respective IC50 values of 24.83 mg/mL and 31.19 mg/mL, respectively. The results reveal that the bio-fabricated ZMC NPs have enhanced cytotoxicity activity more than the standard drug. The result could be due to the synergism of different metals, improved surface-to-volume ratio, and richer electrochemical characteristics [86]. Consequently, bio-prepared ZMC NPs can be a good candidate for developing and designing therapeutic agents mainly for breast cancer ailments.
3.7.2 Antibacterial Activity
The antibacterial efficacy of bio-prepared ZMC NPs against selected pathogenic bacterial strains is presented in Table 3 and Fig. 9. Bio-prepared ZMC NPs have shown substantial antibacterial efficacy against S. aureus (+), S. pneumoniae (+), P. aeruginosa (−), and E. coli (−). At 200 μg/mL ZMC NPs have shown the highest ZOI (35 ± 0.03) with respective MIC of 5 μg/mL for S. aureus strains which slightly preceded the inhibition zone (31 ± 0.11) of chloramphenicol. Comparably, the lowest zone of inhibition was observed (31 ± 0.11with MIC 10 μg/mL) against E. coli with a similar concentration (200 μg/mL). The antibacterial efficacies of ZMC NPs against all tested strains are higher than the standard drug (chloramphenicol). Moreover, the bio-prepared trimetallic ZMC NPs also revealed enhanced antibacterial efficacy than its monometallic counterpart, reported in our previous work [54]. This could be due to the synergistic effects of three metals as well as the modification of physicochemical features including size, surface area, shape, and charge distribution of trimetallic ZMC NPs [87, 88] Therefore, the as-synthesized ZMC NPs can be designed and developed as a drug for the remedies of multidrug-resistant bacterial infections.
3.7.3 Molecular Docking Study
One of the most significant aspects in the design and development of drugs is the evaluation of interaction efficiency and the binding affinity of molecules with particular targets. Using AutoDock 4.2.6 (MGL tools 1.5.7) and a molecular docking database, interactions between bio-prepared ZMC NPs and doxorubicin (standard drug) were examined against the estrogen receptor alpha (ERα; PDB: 5GS4) [62, 89]. Estrogen receptor (ERα) is the primary clinical biomarker employed to subtype cancers of the breast. The estrogen receptor α (ERα) plays an essential role in the progression and development of hormonal-dependent type breast cancer [90]. The molecular docking analysis of ZMC NPs in (Table 4 and Fig. 10a, b) has revealed that two active sites of ERα, Arg363, and Lys449 amino acids which took part in hydrogen bonding. Four amino acid residues, namely Ala322, Glu323, Trp360, and Trp393 involved in van der Waals interactions. Glu353, His356, Met357, and Arg394 are the four amino acid residues participating in π-Sigma/π-Alkyl interaction. On the other hand, doxorubicin in (Table 4 and Fig. 10c, d) has revealed a total of eleven interactions: four hydrogen bonding (Glu353, Arg394, Trp393, Glu323), four π-Sigma/π-Alkyl (Met357, Trp360, Ile386, and His356) interaction and three van der Waals (Gly390, Leu387, and Lys449) interactions.
The binding affinities and inhibition constants of ZMC NPs and doxorubicin were found to be – 9.85 and 1.75, – 7.54 kcal/mol, and 2.99 µM, respectively. The results revealed that both ZMC NPs and the standard drug had shown promising interactions with active sites of amino acid residues of estrogen receptors (ERα). However, the binding affinity of ZMC NPs (– 9.85) kcal/mol) was remarkably higher than the standard drug which is – 7.54 kcal/mol. These demonstrate that the biosynthesized ZMC NPs will have stronger interactions with estrogen receptor alpha (ERα; PDB: 5GS4) than the standard drug. These strong binding with amino acids of estrogen receptors (ERα) is important to prevent the proliferation rates of breast cancer cells [63]. Moreover, the molecular docking interactions are also correlated with the experimental results presented in Table 2, confirming the enhanced cytotoxicity efficacy of ZMC NPs against MCF-7 cell lines. Therefore, bio-prepared ZMC NPs can be promising candidates for breast cancer ailments.
In order to propose the binding mechanism of the bio-prepared nanoparticles and the standard drug (chloramphenicol) with bacterial cells, their interaction efficiency and binding affinity were evaluated against S. aureus dihydrofolate reductase and E. coli DNA gyrase B. Dihydrofolate reductase and DNA gyrase B are the most prominent enzymes for the survival of the respective bacterial species [91]. They are ubiquitously expressed in bacterial species and crucial for cell growth and proliferation. The molecules' interactions with these enzymes are supposed to hinder the DNA replications of this species [92]. Therefore, the good interactions with the amino acid residues of these enzymes could be the antibacterial drug targets of synthesized materials.
The results of molecular docking analysis of ZMC NPs and chloramphenicol (standard drug) against S. aureus have been presented in Table 4, Fig. 11a–d. ZMC NPs has demonstrated binding interaction with amino acids of S. aureus through hydrogen bond (Ser49), π-Sigma/π-Alkyl (Leu28, Val31, Leu54, and Phe92), and van der Waals (Leu20, Ile50, Arg57, and Asp27) interactions with a binding affinity – 12.31 kcal/mol and an inhibition constant (IC) of 9.5 × 10–4 µM. Chloramphenicol has revealed the interactions through hydrogen bond (Trp121, Gln95, and Trp46), and van der Waals (Ile14, Gly93, Gly94, and Ser49) interactions with a respective binding affinity and an IC of – 7.16 kcal/mol and 29.02 µM. respectively. In general, the bioprepared ZMC NPs have strong binding interactions with the prominent bacterial proteins, and that is also associated with the experimental results.
Table 4 and Fig. 12a–d presented the molecular docking assessment of ZMC NPs and chloramphenicol against E. coli DNA gyrase B. ZMC NPs have shown the binding interactions through hydrogen bond (Arg76), π-Sigma/π-Alkyl (Asp49, Ile94, Thr165, and Val167), and van der Waals (Asn46, Glu50, Asp73, and Ile78) interactions with a binding affinity and IC of – 6.04 kcal/mol and an of 37.42 µM, respectively. Chloramphenicol has revealed the interactions through hydrogen bond (Asp73, Asn46, and Glu50), π-Sigma/π-Alkyl (Val43), and van der Waals (Ile78, Arg76, and Trp165) interactions with a binding affinity and an IC of – 6.19 kcal/mol and 5.67 µM, respectively. These results are feasibly complemented with in-vitro activities; hence, the bio-prepared ZMC NPs can be further studied and possibly exploited as antibacterial agents.
4 Conclusions
This study encompasses a one-pot, cost-effective, and environmentally benign synthesis of novel trimetallic ZMC NPs using the medicinal plant A. abyssinica. The SPR peak of the biosynthesized ZMC NPs at 375 nm UV spectrum affirmed the preparations of iso-morphological and spherical nanoparticles. The FTIR and EDX data ascertained the existence of characteristic phytochemicals from AALE accountable for the fabrication of ZMC NPs. The bio-fabricated ZMC NPs have been analyzed by using XRD, FESEM/EDX, TEM/HRTEM/SAED, and the results have revealed a pure, spherical shape with average crystalline and particle sizes of 14.67 and 15.13 nm, respectively. The biological functionalities of bio-fabricated ZMC NPs has been evaluated and revealed enhanced in-vitro cytotoxicity and antibacterial efficacies. Results revealed that the bio-prepared ZMC NPs exhibited high cytotoxicity (94.37 ± 0.14) against MCF-7 cell lines and low cytotoxicity (52.82 ± 0.11) against normal PBMC cell lines by MTT assay. The antibacterial efficacy of ZMC NPs have evaluated against S. pneumoniae, S. aureus, P. aeruginosa, and E. coli strains and has revealed high zones of inhibition such as 35 ± 0.03, 34 ± 0.04, 33 ± 0.14, and 31 ± 0.11 mm, respectively. Furthermore, ZMC NPS also have shown strong interactions with amino acids of estrogen receptor (ERα), S. aureus, and E. coli with binding energies of – 9.85, – 12.31, and – 6.04 kcal/mol, respectively. Consequently, the biosynthesized ZMC NPs have a remarkable cytotoxicity and antibacterial efficacy and can be designed as a therapeutic agent for these ailments. However, further in-vivo biological efficacy studies against MCF-7 cell lines and the selected bacterial strains are recommended.
Data Availability
Data will be made available on request.
References
Yuan P, Ding X, Yang YY, Xu QH (2018) Metal nanoparticles for diagnosis and therapy of bacterial infection. Adv Healthcare Mater 7:1701392
Auría-Soro C, Nesma T, Juanes-Velasco P, Landeira-Viñuela A, Fidalgo-Gomez H et al (2019) Interactions of nanoparticles and biosystems: microenvironment of nanoparticles and biomolecules in nanomedicine. Nanomaterials 9:1365
Medina-Cruz D, Saleh B, Vernet-Crua A, Nieto-Argüello A, Lomelí-Marroquín D, et al. (2020) Bimetallic nanoparticles for biomedical applications: A review. Racing for the Surface: 397–434.
Gao Y, Arokia Vijaya Anand M, Ramachandran V, Karthikkumar V, Shalini V et al (2019) Biofabrication of zinc oxide nanoparticles from Aspergillus niger, their antioxidant, antimicrobial and anticancer activity. J Cluster Sci 30:937–946
Vidhya E, Vijayakumar S, Prathipkumar S, Praseetha P (2020) Green way biosynthesis: Characterization, antimicrobial and anticancer activity of ZnO nanoparticles. Gene Rep 20:100688
Ginting B, Maulana I, Karnila I (2020) Biosynthesis copper nanoparticles using Blumea balsamifera leaf extracts: characterization of its antioxidant and cytotoxicity activities. Surfaces and Interfaces 21:100799
Meer B, Andleeb A, Iqbal J, Ashraf H, Meer K et al (2022) Bio-Assisted Synthesis and Characterization of Zinc Oxide Nanoparticles from Lepidium Sativum and Their Potent Antioxidant. Antibacterial and Anticancer Activities Biomolecules 12:855
Hadi AJ, Nayef UM, Mutlak FA-H, Jabir MS (2023) Laser-ablated zinc oxide nanoparticles and evaluation of their antibacterial and anticancer activity against an ovarian cancer cell line: in vitro study. Plasmonics: 1–11.
Rani N, Rawat K, Saini M, Yadav S, Shrivastava A et al (2022) Azadirachta indica leaf extract mediated biosynthesized rod-shaped zinc oxide nanoparticles for in vitro lung cancer treatment. Mater Sci Eng, B 284:115851
Ahamed M, Lateef R, Khan MM, Rajanahalli P, Akhtar MJ (2023) Biosynthesis, Characterization, and Augmented Anticancer Activity of ZrO2 Doped ZnO/rGO Nanocomposite. Journal of Funct Biomaterials 14:38
Mousa AB, Moawad R, Abdallah Y, Abdel-Rasheed M, Zaher AMA (2023) Zinc Oxide Nanoparticles Promise Anticancer and Antibacterial Activity in Ovarian Cancer. Pharmaceutical Research: 1–10.
Efati Z, Shahangian SS, Darroudi M, Amiri H, Hashemy SI, et al. (2023) Green chemistry synthesized zinc oxide nanoparticles in Lepidium sativum L. seed extract and evaluation of their anticancer activity in human colorectal cancer cells. Ceramics International.
Naser SS, Ghosh B, Simnani FZ, Singh D, Choudhury A et al (2023) Emerging Trends in the Application of Green Synthesized Biocompatible ZnO Nanoparticles for Translational Paradigm in Cancer Therapy. Journal of Nanotheranostics 4:248–279
Aboul-Soud MA, Siddique R, Fozia F, Ullah A, Rashid Y, et al. (2023) Antiplatelet, cytotoxic activities and characterization of green-synthesized zinc oxide nanoparticles using aqueous extract of Nephrolepis exaltata. Environmental Science and Pollution Research: 1–11.
Dolati M, Tafvizi F, Salehipour M, Komeili Movahed T, Jafari P (2023) Biogenic copper oxide nanoparticles from Bacillus coagulans induced reactive oxygen species generation and apoptotic and anti-metastatic activities in breast cancer cells. Sci Rep 13:3256
Younas M, Zubair M, Rizwan M, Khan MA, Hussaini KM et al (2023) Synthesis and characterization of cerium, silver and copper oxide nanoparticles and their anticancer potential of hepatocellular carcinoma HepG2 cancer cells. J Mol Struct 1288:135756
Doman KM, Gharieb MM, Abd El-Monem AM, Morsi HH (2023) Synthesis of silver and copper nanoparticle using Spirulina platensis and evaluation of their anticancer activity. International Journal of Environmental Health Research: 1–13.
Mohammad MS, Shyam P (2023) Structural Analysis of Biogenic Copper Oxide Nanoparticles, and their Bio-Activity Assessment. BioNanoScience: 1–11.
Sathiyavimal S, Vasantharaj S, Kaliannan T, Garalleh HA, Garaleh M et al (2023) Bio-functionalized copper oxide/chitosan nanocomposite using Sida cordifolia and their efficient properties of antibacterial, anticancer activity against on breast and lung cancer cell lines. Environ Res 218:114986
Fathy RM, Mahfouz AY (2021) Eco-friendly graphene oxide-based magnesium oxide nanocomposite synthesis using fungal fermented by-products and gamma rays for outstanding antimicrobial, antioxidant, and anticancer activities. Journal of Nanostructure in Chemistry 11:301–321
Ali S, Sudha KG, Thirumalaivasan N, Ahamed M, Pandiaraj S et al (2023) Green Synthesis of Magnesium Oxide Nanoparticles by Using Abrus precatorius Bark Extract and Their Photocatalytic, Antioxidant, Antibacterial, and Cytotoxicity Activities. Bioengineering 10:302
Fouda A, Eid AM, Abdel-Rahman MA, El-Belely EF, Awad MA et al (2022) Enhanced antimicrobial, cytotoxicity, larvicidal, and repellence activities of brown algae, Cystoseira crinita-mediated green synthesis of magnesium oxide nanoparticles. Frontiers in Bioengineering and Biotechnology 10:849921
Abdelghany TM, Al-Rajhi AM, Yahya R, Bakri MM, Al Abboud MA et al (2023) Phytofabrication of zinc oxide nanoparticles with advanced characterization and its antioxidant, anticancer, and antimicrobial activity against pathogenic microorganisms. Biomass Conversion and Biorefinery 13:417–430
Pugazhendhi A, Prabhu R, Muruganantham K, Shanmuganathan R, Natarajan S (2019) Anticancer, antimicrobial and photocatalytic activities of green synthesized magnesium oxide nanoparticles (MgONPs) using aqueous extract of Sargassum wightii. J Photochem Photobiol, B 190:86–97
Sathiyavimal S, Durán-Lara EF, Vasantharaj S, Saravanan M, Sabour A et al (2022) Green synthesis of copper oxide nanoparticles using Abutilon indicum leaves extract and their evaluation of antibacterial, anticancer in human A549 lung and MDA-MB-231 breast cancer cells. Food Chem Toxicol 168:113330
Basavegowda N, Baek K-H (2021) Multimetallic nanoparticles as alternative antimicrobial agents: challenges and perspectives. Molecules 26:912
Abid S, Uzair B, Niazi MBK, Fasim F, Bano SA et al (2021) Bursting the virulence traits of MDR strain of Candida albicans using sodium alginate-based microspheres containing nystatin-loaded MgO/CuO nanocomposites. Int J Nanomed 16:1157
Cao Y, Dhahad HA, El-Shorbagy M, Alijani HQ, Zakeri M et al (2021) Green synthesis of bimetallic ZnO–CuO nanoparticles and their cytotoxicity properties. Sci Rep 11:23479
Habibullah G, Viktorova J, Ruml T (2021) C urrent Strategies for Noble Metal Nanoparticle Synthesis. Nanoscale Res Lett 16:1–12
Jiang X, Fan X, Xu W, Zhang R, Wu G (2019) Biosynthesis of bimetallic Au–Ag nanoparticles using Escherichia coli and its biomedical applications. ACS Biomater Sci Eng 6:680–689
Roopan SM, Surendra TV, Elango G, Kumar SHS (2014) Biosynthetic trends and future aspects of bimetallic nanoparticles and its medicinal applications. Appl Microbiol Biotechnol 98:5289–5300
Khan AU, Khan QU, Tahir K, Ullah S, Arooj A et al (2021) A Tagetes minuta based eco-benign synthesis of multifunctional Au/MgO nanocomposite with enhanced photocatalytic, antibacterial and DPPH scavenging activities. Mater Sci Eng, C 126:112146
Parveen K, Banse V, Ledwani L. Green synthesis of nanoparticles: their advantages and disadvantages; 2016. AIP Publishing LLC. pp. 020048.
Kashid Y, Ghotekar S, Bilal M, Pansambal S, Oza R, et al. (2022) Bio-inspired sustainable synthesis of silver chloride nanoparticles and their prominent applications. Journal of the Indian Chemical Society: 100335.
Ghotekar S, Pansambal S, Bilal M, Pingale SS, Oza R (2021) Environmentally friendly synthesis of Cr2O3 nanoparticles: characterization, applications and future perspective─ a review. Case Studies in Chemical and Environmental Engineering 3:100089
Dauthal P, Mukhopadhyay M (2016) Noble metal nanoparticles: plant-mediated synthesis, mechanistic aspects of synthesis, and applications. Ind Eng Chem Res 55:9557–9577
Khatami M, Varma RS, Zafarnia N, Yaghoobi H, Sarani M et al (2018) Applications of green synthesized Ag, ZnO and Ag/ZnO nanoparticles for making clinical antimicrobial wound-healing bandages. Sustainable Chemistry and Pharmacy 10:9–15
Tiri RNE, Gulbagca F, Aygun A, Cherif A, Sen F (2022) Biosynthesis of Ag–Pt bimetallic nanoparticles using propolis extract: Antibacterial effects and catalytic activity on NaBH4 hydrolysis. Environ Res 206:112622
Aygun A, Gulbagca F, Altuner EE, Bekmezci M, Gur T, et al. (2022) Highly active PdPt bimetallic nanoparticles synthesized by one-step bioreduction method: Characterizations, anticancer, antibacterial activities and evaluation of their catalytic effect for hydrogen generation. International Journal of Hydrogen Energy.
Barui AK, Das S, Patra CR (2019) Biomedical applications of green-synthesized metal nanoparticles using polysaccharides. Elsevier, Functional Polysaccharides for Biomedical Applications, pp 329–355
Majeed M, Hakeem KR, Rehman RU (2022) Synergistic effect of plant extract coupled silver nanoparticles in various therapeutic applications-present insights and bottlenecks. Chemosphere 288:132527
Deo S, Sharma J, Kumar S (2022) GLOBOCAN 2020 report on global cancer burden: challenges and opportunities for surgical oncologists. Ann Surg Oncol 29:6497–6500
Ikuta KS, Swetschinski LR, Aguilar GR, Sharara F, Mestrovic T et al (2022) Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet 400:2221–2248
Findlater A, Bogoch II (2018) Human mobility and the global spread of infectious diseases: a focus on air travel. Trends Parasitol 34:772–783
Bayón-Cordero L, Alkorta I, Arana L (2019) Application of solid lipid nanoparticles to improve the efficiency of anticancer drugs. Nanomaterials 9:474
Rajani C, Borisa P, Karanwad T, Borade Y, Patel V et al (2020) Cancer-targeted chemotherapy: emerging role of the folate anchored dendrimer as drug delivery nanocarrier. Elsevier, Pharmaceutical applications of dendrimers, pp 151–198
Michael GB, Freitag C, Wendlandt S, Eidam C, Feßler AT et al (2015) Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol 10:427–443
Cucci M, Wooten C, Fowler M, Mallat A, Hieb N et al (2020) Incidence and Risk Factors Associated with Multi-Drug–Resistant Pathogens in a Critically Ill Trauma Population: A Retrospective Cohort Study. Surg Infect 21:15–22
Asfaw N, Demissew S (2015) Essential oil composition of four Artemisia Species from Ethiopia. Bull Chem Soc Ethiop 29:123–128
Tesfahuneygn G, Gebreegziabher G (2019) Medicinal plants used in traditional medicine by Ethiopians: a review article. J Respir Med Lung Dis 4:1–3
d’Avigdor E, Wohlmuth H, Asfaw Z, Awas T (2014) The current status of knowledge of herbal medicine and medicinal plants in Fiche. Ethiopia Journal of ethnobiology and ethnomedicine 10:38
Ayalew H, Tewelde E, Abebe B, Alebachew Y, Tadesse S (2022) Endemic medicinal plants of Ethiopia: Ethnomedicinal uses, biological activities and chemical constituents. J Ethnopharmacol 293:115307
Adugna M, Feyera T, Taddese W, Admasu P (2014) In vivo antimalarial activity of crude extract of aerial part of Artemisia abyssinica against Plasmodium berghei in mice. Global J Pharmacol 8:460–468
Achamo T, Zereffa EA, Murthy HA, Ramachandran VP, Balachandran R (2022) Phyto-mediated synthesis of copper oxide nanoparticles using Artemisia abyssinica leaf extract and its antioxidant, antimicrobial and DNA binding activities. Green Chem Lett Rev 15:598–614
Letha N, Ganesan K, Nair SKP, Gani S (2016) Studies on phytochemical screening and in vitro antioxidant activity of Ethiopian indigenous medicinal Plants, Artemisia abyssinica Sch. Bip. ex A. Rich World J Pharm Res 5:1048–1058
Aman M, Asfaw Z, Dalle G (2020) Medicinal Plants Used for Treating Human and Livestock Ailments in Tiyo District, Arsi Zone of Oromia, Ethiopia.
Elemike EE, Onwudiwe DC, Nundkumar N, Singh M, Iyekowa O (2019) Green synthesis of Ag, Au and Ag-Au bimetallic nanoparticles using Stigmaphyllon ovatum leaf extract and their in vitro anticancer potential. Mater Lett 243:148–152
Dekkers JF, van Vliet EJ, Sachs N, Rosenbluth JM, Kopper O et al (2021) Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat Protoc 16:1936–1965
Rai Y, Pathak R, Kumari N, Sah DK, Pandey S et al (2018) Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci Rep 8:1–15
Nga N, Ngoc T, Trinh N, Thuoc T, Thao D (2020) Optimization and application of MTT assay in determining density of suspension cells. Anal Biochem 610:113937
Yaqub A, Malkani N, Shabbir A, Ditta SA, Tanvir F et al (2020) Novel biosynthesis of copper nanoparticles using Zingiber and Allium sp. with synergic effect of doxycycline for anticancer and bactericidal activity. Curr Microbiol 77:2287–2299
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK et al (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Khalil AT, Ayaz M, Ovais M, Wadood A, Ali M et al (2018) In vitro cholinesterase enzymes inhibitory potential and in silico molecular docking studies of biogenic metal oxides nanoparticles. Inorganic and Nano-Metal Chemistry 48:441–448
Saghir AV, Beidokhti SM, Khaki JV, Salimi A (2021) One-step synthesis of single-phase (Co, Mg, Ni, Cu, Zn) O High entropy oxide nanoparticles through SCS procedure: Thermodynamics and experimental evaluation. J Eur Ceram Soc 41:563–579
Pandey KB, Rizvi SI (2009) Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev 2:270–278
Al Mamari HH (2021) Phenolic compounds: Classification, chemistry, and updated techniques of analysis and synthesis. Phenolic compounds-chemistry, synthesis, diversity, non-conventional industrial, pharmaceutical and therapeutic applications 10.
Thamer N, Muftin N, Al-Rubae S (2018) Optimization properties and characterization of green synthesis of copper oxide nanoparticles using aqueous extract of Cordia myxa L. leaves. Asian J Chem 30:1559–1563
Annathurai S, Chidambaram S, Baskaran B, Prasanna Venkatesan G (2019) Green synthesis and electrical properties of p-CuO/n-ZnO heterojunction diodes. J Inorg Organomet Polym Mater 29:535–540
Majhi JK, Kuiri PK (2021) Large spectral shift of plasmon resonances in Au–Cu alloy nanoparticles through anisotropy and interaction. Bull Mater Sci 44:18
Mlalila NG, Swai HS, Hilonga A, Kadam DM (2016) Antimicrobial dependence of silver nanoparticles on surface plasmon resonance bands against Escherichia coli. Nanotechnology, science and applications: 1–9.
Vinu D, Govindaraju K, Vasantharaja R, Amreen Nisa S, Kannan M et al (2021) Biogenic zinc oxide, copper oxide and selenium nanoparticles: preparation, characterization and their anti-bacterial activity against Vibrio parahaemolyticus. Journal of Nanostructure in Chemistry 11:271–286
Ajitha B, Reddy YAK, Reddy PS, Jeon H-J, Ahn CW (2016) Role of capping agents in controlling silver nanoparticles size, antibacterial activity and potential application as optical hydrogen peroxide sensor. RSC Adv 6:36171–36179
Khani R, Roostaei B, Bagherzade G, Moudi M (2018) Green synthesis of copper nanoparticles by fruit extract of Ziziphus spina-christi (L.) Willd.: application for adsorption of triphenylmethane dye and antibacterial assay. J Mol Liq 255:541–549
Rajagopal G, Nivetha A, Sundar M, Panneerselvam T, Murugesan S et al (2021) Mixed phytochemicals mediated synthesis of copper nanoparticles for anticancer and larvicidal applications. Heliyon 7:e07360
Sarwar N, Humayoun UB, Kumar M, Zaidi SFA, Yoo JH et al (2021) Citric acid mediated green synthesis of copper nanoparticles using cinnamon bark extract and its multifaceted applications. J Clean Prod 292:125974
Umaralikhan L, Jamal Mohamed Jaffar M (2018) Green synthesis of MgO nanoparticles and it antibacterial activity. Iranian Journal of Science and Technology, Transactions A: Science 42:477–485
Lv Q, Zhang B, Xing X, Zhao Y, Cai R et al (2018) Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: Novel approach and mechanisms investigation. J Hazard Mater 347:141–149
Labhane P, Huse V, Patle L, Chaudhari A, Sonawane G (2015) Synthesis of Cu doped ZnO nanoparticles: crystallographic, optical, FTIR, morphological and photocatalytic study. Journal of Materials Science and Chemical Engineering 3:39
Pillai AM, Sivasankarapillai VS, Rahdar A, Joseph J, Sadeghfar F et al (2020) Green synthesis and characterization of zinc oxide nanoparticles with antibacterial and antifungal activity. J Mol Struct 1211:128107
Singh A, Gaud B, Jaybhaye S (2020) Optimization of synthesis parameters of silver nanoparticles and its antimicrobial activity. Materials Science for Energy Technologies 3:232–236
Noori AJ, Kareem FA (2019) The effect of magnesium oxide nanoparticles on the antibacterial and antibiofilm properties of glass-ionomer cement. Heliyon 5:e02568
Wong CW, Chan YS, Jeevanandam J, Pal K, Bechelany M et al (2020) Response surface methodology optimization of mono-dispersed MgO nanoparticles fabricated by ultrasonic-assisted sol–gel method for outstanding antimicrobial and antibiofilm activities. J Cluster Sci 31:367–389
Maheo AR, Vithiya BSM, Arul Prasad TA, Mangesh V, Perumal T et al (2023) Cytotoxic, Antidiabetic, and Antioxidant Study of Biogenically Improvised Elsholtzia blanda and Chitosan-Assisted Zinc Oxide Nanoparticles. ACS Omega 8:10954–10967
Khonkarn R, Okonogi S, Ampasavate C, Anuchapreeda S (2010) Investigation of fruit peel extracts as sources for compounds with antioxidant and antiproliferative activities against human cell lines. Food Chem Toxicol 48:2122–2129
Ioset J-R, Brun R, Wenzler T, Kaiser M, Yardley V (2009) Drug screening for kinetoplastids diseases. A training manual for screening in neglected diseases.
Zughaibi TA, Mirza AA, Suhail M, Jabir NR, Zaidi SK, et al. (2022) Evaluation of anticancer potential of biogenic copper oxide nanoparticles (CuO NPs) against breast cancer. Journal of Nanomaterials 2022.
Nieto-Maldonado A, Bustos-Guadarrama S, Espinoza-Gomez H, Flores-López LZ, Ramirez-Acosta K, et al. (2022) Green synthesis of copper nanoparticles using different plant extracts and their antibacterial activity. Journal of Environmental Chemical Engineering: 107130.
Gupta MN, Khare SK, Sinha R (2021) Interfaces Between Nanomaterials and Microbes: CRC Press.
Anza M, Endale M, Cardona L, Cortes D, Eswaramoorthy R et al (2021) Cytotoxicity, antimicrobial activity, molecular docking, drug likeness and DFT analysis of benzo [c] phenanthridine alkaloids from roots of Zanthoxylum chalybeum. Biointerface Research in Applied Chemistry 12:1569–1586
Bhatt S, Stender J, Joshi S, Wu G, Katzenellenbogen B (2016) OCT-4: A novel estrogen receptor-α collaborator that promotes tamoxifen resistance in breast cancer cells. Oncogene 35:5722–5734
Srinivasan B, Tonddast-Navaei S, Roy § A, Zhou H, Skolnick J, (2019) Chemical space of Escherichia coli dihydrofolate reductase inhibitors: New approaches for discovering novel drugs for old bugs. Med Res Rev 39:684–705
Bhattacharya S, Sen D, Bhattacharjee C (2022) Inhibition mechanism study for diallyl thiosulfinate (allicin) against crucial bacterial proteins through in silico molecular docking simulation. Process Biochem 122:110–119
Acknowledgements
The authors are grateful for material and technical assistance from Haramaya University, Adama Science and Technology University, and University of Botswana.
Funding
Open access funding provided by University of Botswana.
Author information
Authors and Affiliations
Contributions
TAO: Data curation, conceptualization, formal analysis, methodology, investigation, writing- original draft preparation; EAZ: Conceptualization, investigation, methodology, validation, supervision, writing- reviewing and editing; HCAM: Conceptualization, investigation, methodology, validation, supervision, writing reviewing and editing; TBD: Investigation, methodology, analysis, software, validations, editing and reviewing; OP: Analysis, software, validations, editing and reviewing; KP: Analysis, software, validations, editing and reviewing; SG: Analysis, software, validations, editing and reviewing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Orshiso, T.A., Zereffa, E.A., Murthy, H.C.A. et al. One-Pot Biopreparation of Trimetallic ZnO–MgO–CuO Nanoparticles: Enhanced Cytotoxicity, Antibacterial Activities and Molecular Docking Studies. Chemistry Africa 7, 1963–1980 (2024). https://doi.org/10.1007/s42250-023-00830-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-023-00830-0