Abstract
Introduction
Theaflavins belong to the class of polyphenols that are predominantly found in black tea. The major derivatives of theaflavins found in black tea are theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B), and theaflavin-3,3′-digallate (TF3). Theaflavin-3,3′-digallate (TF3) is a natural compound present in black tea and known to possess antiviral activity. This study had attempted to explore the potential role of TF3 in inhibiting various stages of the SARS-CoV-2 life cycle.
Methods
Molecular docking studies of TF3 along with positive controls was performed on eight different targets of SARS-CoV-2 followed by binding free energy (MM-GBSA) calculations. The docked complexes with favourable docking and binding free energy results were subjected to molecular dynamics (MD) simulation studies to assess the stability of the dock complex. Finally, TF3 and all the positive controls were taken for ADMET analysis.
Results
The docking and binding free energy results of TF3 was compared against the positive controls. TF3 showed the highest binding energy against all the targets and formed more stable interactions for a longer duration on MD simulations with CLpro, RdRp, helicase and spike protein. Also, the promising in-silico ADMET profile further warrants the exploration of this compound through in-vitro and in-vivo methods.
Conclusion
Through this study, we tried to evaluate the role of theaflavin-3,3’-digallate on multiple targets of SARS-CoV-2, and the positive in-silico results which were obtained on various pharmacodynamic and pharmacokinetic parameters, give a ray of hope as a potential therapeutic drug to this rapidly spreading disease. The search for a curative therapy for SARS-CoV-2 is still ongoing. The favourable preliminary results of TF3 through in-silico analysis offers a ray of hope in ending this devasting pandemic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
SARS-CoV-2 belongs to the family of coronaviridae whose genomic structure is closely related to severe acute respiratory syndrome (SARS-CoV) and middle-east respiratory syndrome coronavirus (MERS-CoV). In comparison with both (i.e., SARS-CoV and MERS-CoV), the novel coronavirus has a higher transmission rate. Due to the higher transmission rate and death cases, it was declared a global pandemic by World Health Organization (WHO) in March 2020 [1, 2]. The literature data suggest that the novel coronavirus spike protein has a potent binding affinity with human angiotensin-converting enzyme 2 (ACE2) receptor present on the cell surface and this evidence may explain that, why these species have a higher transmission rate as compared to other corona viruses [3,4,5,6,7].
SARS-CoV-2 is a type of RNA virus that is about 30 kb in size, encoded with various structural and non-structural proteins. The structural proteins have four different types- spike protein (S), membrane (M), envelope (E), and nucleocapsid (N), which occupies 33% of the viral genome at the C-terminus and play an important role in viral integrity. The non-structural proteins are divided into sixteen different types, which are important for various stages of the viral life cycle [8, 9]. To date, many studies have been done to evaluate the molecular mechanism involved in the SARS-CoV-2 life cycle and to identify the targets involved in it. The possible therapeutic strategies to target viruses can divide into four types that aim to target cellular machinery at different stages of the viral life cycle (Fig. 1).
The life cycle of SARS-CoV-2 and potential therapeutic targets: The viral life cycle has 4 stages—entry, replication, the release of viral progenies, and affecting the RAAS system. The inhibition of 1. spike protein and ACE2 interaction, 2. TMPRSS2 activity, which mediates the cleavage of spike protein, and 3. clathrin-mediated endocytosis prevents the entry of virus in the cell. Inhibition of 4. CLpro and PLpro, to viral proteases, inhibit the multiplication of virus while targeting 5. RdRp and helicase or 6. increasing intracellular Zn + 2 concentration inhibit replication. 7. decreasing expression and activity of veroporin3, an ion channel prevents the release of virions and thus infection in other cells. RAAS overactivity can be suppressed by inhibiting 8. ACE and 9. AT1R 3CLpro- chymotrypsin-like protease. PLpro papain-like protease, ACE angiotensin-converting enzyme, AT1R angiotensin II type 1 receptor, Ang angiotensin, MasR mitochondrial assembly receptor, M membrane, S spike, E envelope, N nucleocapsid, TMPRSS2 transmembrane protease serine 2, RdRp RNA dependant RNA polymerase, RTC replication transcription complex, RAAS renin–angiotensin–aldosterone system, pp polyprotein
The first therapeutic strategy is to block the entry of the virus. This could be achieved by blockage of proteins that promote viral entry and fusion into the host cell. Viral entry into the human cell is initiated through the interaction between viral spike protein (S) and angiotensin-converting enzyme2 (ACE2) receptor at receptor binding domain. Upon binding with ACE2, spike protein undergoes conformational change followed by cleavage between S1 and S2 domain. Initially, furin followed by Transmembrane Serine Protease 2 (TMPRSS2) involved in the cleavage. S1 helps in binding with host cell via receptor binding domain whereas S2 promotes the endocytic entry of virus by membrane fusion that completes the infection process [10,11,12].
The second and third strategies are aimed at targeting proteins that are involved in viral replication and release [13]. Upon membrane fusion, the virus releases single-stranded RNA that goes under the translation process and cleaved into two precursor polyproteins pp1a and pp1b. Both pp1a and pp1b are further cleaved by viral protease into non-structural proteins. The non-structural proteins are divided into sixteen different types that include important replicating enzymes like RNA-dependent RNA polymerase (RdRp, nsp12), helicase (nsp13), papain-like protease (PLpro, nsp3), chymotrypsin-like main protease (3CL protease, nsp5), and exo-ribonuclease (nsp14). These non-structural proteins form a replication transcription complex (RTC) that undergoes transcription and translation to synthesize full-length genomic RNA (transcription) or nested subgenomic mRNA (translation) which is further translated into structural proteins. The structural protein in association with the viral genome is assembled into new virions and finally released through the budding process [14, 15].
The fourth strategy involved in modulation of the immune system with RAAS. RAAS is involved in a variety of biological responses that include regulation of inflammation, blood pressure, and fibrosis. In general angiotensin converting enzyme convert angiotensin I to angiotensin II. By ACE2 enzyme, angiotensin II is further converted into angiotensin (1–7) and binds with G-protein coupled receptor Mas to reduce inflammation, blood pressure, and fibrosis, thereby providing a protective effect in the lungs. Upon binding of SARS-CoV-2 with ACE2; the normal function of ACE2 is suppressed and instead of conversion into angiotensin (1–7); ACE2 binds with the AT1R receptor which causes an increase in inflammation, blood pressure, and fibrosis [16]. As the RAAS system is involved in a variety of functions, targeting the RAAS system needs more specificity [17].
Theaflavins belong to the class of polyphenols that are predominantly found in black tea. The major derivatives of theaflavins found in black tea are theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B), and theaflavin-3,3′-digallate (TF3). All of the derivatives have been reported for a variety of biological activities like anti-viral, anti-tumour, anti-oxidant, and antibacterial activities [18, 19]. Literature data suggest that theaflavin derivatives found in black tea had a wide spectrum of anti-viral activity that can act at different stages of the viral life cycle [20]. Since the theaflavins obtained from the natural source, anti-viral promising results in past encouraged us to explore the role of theaflavin-3,3′-digallate in to fight against SARS-CoV-2 using the in-silico approach. In our study, we have included various druggable targets of SARS-CoV-2 which are Chymotrypsin-like protease (3CLpro), RNA dependant RNA polymerase (RdRp), Papain like protease (PLpro), Helicase, Spike Receptor Binding Domain (RBD), endoribonuclease, and the human targets like furin and TMPRSS2 involved in binding with the virus and evaluated the potential role of theaflavin-3,3′-digallate against SARS-CoV-2 using in-silico approach. The insilico methods includes molecular docking studies and MD simulation studies on eight different druggable targets of SARS-CoV-2; with this we had also included insilico ADME/T studies.
2 Material and Methods
2.1 Preparation of Ligands
We downloaded the structures of theaflavin-3,3′-digallate (21,146,795 CID, Mol. Wt. 868.7), remdesivir from Pubchem (121304016 CID), and extracted cognate ligands (positive controls) (Fig. 2) from the crystal structure of the protein downloaded from Protein Data Bank (https://www.rcsb.org/). Additionally, we used a reported inhibitor of TMPRSS2, camostat (PubChem CID 2536) [21]. Ligand preparation was done using Schrodinger’s LigPrep module (Schrödinger, LLC, New York, NY, 2020). All possible states at target pH 7 ± 2 were generated using the Epik tool [22]. Tautomers were also generated by retaining specific chiralities and varying other chiral centres. Energy minimization was done using the OPLS3 force field [23].
2.2 Protein Structure Retrieval
For this study, we retrieved the structures from the protein data bank in the PDB format for all the targets except TMPRSS2. The PDB ID of all proteins is mentioned in table no 1. For TMPRSS2, we used the previously reported modelled structure [24]. We further refine the structure using the galaxy refine server [25] and validated it through the Molprobidity server [26] and Schrodinger’s structure analysis tool.
2.3 Protein Preparation
The protein Preparation Wizard tool [27] of Maestro software from Schrodinger, LLC, 2020 was used to prepare the protein structures before molecular docking studies. The structures were pre-processed and H-bond assignment, geometry optimization, and energy minimization of protein were done at physiological pH with OPLS3 (Optimized Potential for Liquid Simulations version 3.0) force field. Water molecules present in the active sites of CLpro and Plpro were retained. In the preparation of endoribonuclease protein, we removed Chain B and all other Het-atoms as it is a homodimer and both the chains are having same function so using a single chain will make the computational process less intensive. Het-atoms are removed as they are not involved in the binding process and removing them will make the calculations easy. For spike protein preparation, the ACE2 protein chain was removed from the complex. The prime module [28] was used to fill missing residues in RdRp protein preparation.
2.4 Active Site Selection and Receptor Grid Generation
Active site selection and receptor grid generation (Table 1) was done based on the interacting residues of the cognate ligands present with the protein structures except in Spike protein, TMPRSS2, and Helicase. For Spike protein, interacting residues of RBD of Spike protein with ACE2 were selected for the active site. For helicase, interacting residues of Helicase (nsp13) with nsp7-nsp8-nsp12 complex were chosen [29]. For TMPRSS2, interacting residues between TMPRSS2 and SARS CoV 2 Spike glycoprotein were chosen as the active site [24].
2.5 Molecular Docking
For molecular docking studies, the LigPrep file and glide receptor grid file of each target was used as input. The ligand docking was done using Glide extra precision (XP) model [30] with OPLS3 force field [23]. The out file was generated and viewed in a pose viewer tool.
2.6 Binding Energy Calculation
The MM-GBSA binding free energy (ΔGbind) [31] was calculated using pose viewer file generated after molecular docking using Prime module of Schrodinger, LLC, 2020 which defines binding energy in the algorithm as:
2.7 Molecular Dynamic (MD) Simulations
MD simulation was done through the Desmond simulation package (D. E. Shaw Research, New York, NY, 2020). We ran the MD simulation for all the protein targets with TF3, and their positive controls. Additionally, for RdRp, we ran MD simulation with remdesivir also. Protein–ligand complex was selected for MD study based on best binding free energy (MM-GBSA). For system-building, the Transferable Intermolecular Interaction Potential 3 Points (TIP3P) [32] solvation model was chosen with an orthorhombic box of 10 × 10x10 Å. The system was made electrically neutral by adding counter ions (Na+ or Cl−). Salt (NaCl) at the concentration of 0.15 M was added to mimic physiological conditions. The model system was relaxed before the simulation. The OPLS3 force field was used for MD simulation. NPT ensemble with a temperature of 310 K and a pressure of 1 atm was applied during all MD simulations. For Coulombic interactions, 9 Å was chosen as a cut-off radius. Langevin thermostat (relaxation time 1 ps) and barostat (relaxation time 2 ps) [33] were chosen to control temperature and pressure respectively. To calculate non-bonded forces, the RESPA integrator was used and recording was done at every 2 fs. The simulation was done for each protein–ligand complex for 20 ns. The trajectories were saved at every 10 ps and to evaluate the stability of the protein–ligand complexes, the root means square deviation (RMSD) of both the protein and the ligands was computed and inspected. To analyze the local fluctuations in the structures, we also calculated root mean square fluctuations (RMSF) for all the targets. MD results were extracted and viewed using the simulation interaction diagram tool.
2.8 ADMET Studies
Prediction of ADMET properties is one of the important parameters in computer-aided drug discovery to screen the molecules. To evaluate the ADMET profile of theaflavin-3,3′-digallate, remdesivir, and cognate ligand we used the pkCSM online server (http://biosig.unimelb.edu.au/pkcsm/prediction). The smiles of all structures were collected and used as input to predict ADMET properties.
3 Results
3.1 Molecular Docking Analysis
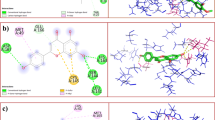
Molecular docking studies were performed to evaluate the binding affinity and to identify molecular interactions between ligand and target. In our study, we had performed molecular docking studies on eight different targets of SARS-CoV-2. All of the receptors which we had included in the study are known to involve in various stages of the life cycle of SARS-CoV-2. The results of molecular docking studies of TF3 were compared with the cognate ligand present with the protein structure for 3CLpro, PLpro, endoribonuclease and furin (Table 2). For RdRp, the results of TF3 were compared with its cognate ligand and remdesivir. For helicase, spike protein and TMPRSS2, the molecular docking studies were performed only with TF3. In addition to this, each ligand–protein complex was analyzed through H-bonded and non-bonded interactions (Fig. 3 and Supplementary S2). The binding affinity of the protein–ligand complex is due to various bonded and non-bonded interactions, in that H-bond interactions play a vital role in the stability of the docked complex. CLpro and furin showed the maximum no. H-bonded interactions with TF3 (10), followed by RdRp (8), endoribonuclease (7), TMPRSS2 and helicase (each 6), spike protein (4) and PLpro (2). The interacting residues for both bonded and non-bonded interactions had also been reported (Table 3).
3.2 Binding Free Energy Analysis (MM-GBSA)
While the docking score measures the binding affinity of a ligand to the protein at a single time point for a given pose, the docking results may not accurately predict the correct binding pose alone [34]. Binding free energy shows which protein–ligand complex has the lowest energy. Combining the docking and MM-GBSA methods can result in the better selection of the ligands to be taken for further studies as it markedly improves the probability of finding the correct binding pose depending upon the interactions of interest between the ligand and the protein and lowest energy of the complex [35,36,37]. Hence, for MD studies we chose the complex for each ligand based on their docking score and the lowest binding free energy as depicted by MM-GBSA calculations. TF3 showed the highest binding free energy with all the targets (Table 2) and thus can show better interactions than the cognate ligand with multiple targets.
3.3 Molecular Dynamics (MD) Analysis
MD studies are performed to understand the stability of protein–ligand complex and the binding interactions between them with reference to time in a simulated physiological condition. For each of the protein–ligand complex, we calculated RMSD values of protein and ligand in complex, RMSF values of the protein, and protein–ligand contact mapping. RMSD values measure the average variations in the displacement of the protein–ligand complex in respect to the initial frame (docked complex which was chosen as an input file for MD). RMSF values measure local fluctuations in the amino acid residues upon ligand binding and it is important to characterize its effect on individual residues of the protein. Since interactions play an important role in both pharmacokinetics and pharmacodynamics, we extracted and mapped various intermolecular interactions like H-bond and other non-bonded interactions from MD trajectory for each of the protein–ligand complex.
During MD studies, the stability of a complex is indicated by how quickly the protein, as well as ligand, attained equilibrium. In a protein-TF3 complex, 3CLpro reached and attained the equilibrium fastest (~ 2.5 ns), while all other proteins attained the same within 5-6 ns. Also, the RMSD values in the protein-TF3 complex remained well within 2–3 Å for all the proteins except for TMPRSS2, where RMSD values were ~ 8–9 Å. When comparison was made between protein-TF3 and protein-cognate/positive control complexes for each target, all the proteins achieved stability faster when complexed with TF3 than cognate ligand/positive controls except with furin. RMSD fluctuations for each target protein were almost the same when complexed with TF3 and its cognate/positive control. TF3 showed the least RMSD fluctuations in 3CLpro complex and TMPRSS2 (2–5 Å) followed by the furin (4 Å) spike (3–7 Å) and RdRp (4–7 Å), endoribonuclease (5–7 Å), PLpro (7–8 Å) and maximum fluctuations in helicase (8–9 Å). Cognate ligand/positive controls showed less or almost RMSD values than TF3 in RdRp (suramin 1–6 Å), PLpro (~ 4 Å) furin (3 Å), and TMPRSS2 (4 Å), while a higher value in helicase (8–9 Å), RdRp (remdesivir 7 Å), endoribonuclease (3–7 Å) and 3CLpro (5 Å). Individual amino acids residues in each protein have RMSF values of < 3 Å in complexes with TF3 and cognate/positive ligand. Combining all the above MD study results, it can be concluded that TF3 formed better and stable interactions with CLpro, RdRp, spike protein and endoribonuclease (Figs. 4, 5, 6, 7). While cognate ligand showed better interactions with PLpro (Supplementary S2) and furin (Supplementary S2). For helicase (Supplementary S2) TF3 fluctuations were very higher and results remained indeterminate with TMPRSS2 (Supplementary S2).
3.4 ADMET Studies
The ADMET properties play a crucial role in the development of new drugs. The in-silico ADMET studies are one of the important techniques that reduce the chances of drug molecules to be failed in further pre-clinical and clinical studies. We had performed in-silico ADMET studies of theaflavin-3,3′-digallate and all the positive controls using the pkCSM webserver. An ideal drug molecule should have good intestinal absorption property, Log S should in between − 1 and − 5, should be a non-inhibitor of CYP450 and should be non-AMES toxic. Apart from this it should be non-carcinogenic, non-inhibitors of hERG and should have less toxicity. ADMET prediction of TF3 showed acceptable values of Log S (solubility), an important parameter that affects in-vitro assays, absorption, bioavailability and formulation. TF3 showed poor intestinal absorption property as well as poor BBB and CNS permeability. While poor absorption can be a limiting factor in an oral formulation, poor CNS permeability ensures that it is devoid of any CNS side effects. The predicted metabolism data suggested that TF3 is a non-inhibitor of the CYP450 enzyme family which indicates that the compound may have good metabolism in the liver and lower chances of drug-drug interactions. The toxicity profile of TF3 showed that it doesn’t have carcinogenic potential (AMES negativity), no cardiotoxicity (non-inhibition of hERG channels) and no hepatotoxicity. The AMES test predicts the ability of a molecule to a mutation in DNA, while blockade of hERG channels shows the potential of causing QT syndrome and sudden deaths. All the prediction on the in-silico ADMET profile suggests that TF3 may have promising ADMET profile that is suitable for a drug molecule.
4 Discussion
The first evidence of SARS-CoV-2 was traced back to December 2019 [38] and even till now, there is no specific therapy available to treat this disease. Many therapeutic options including small molecules as well as vaccines are already in different phases of clinical trials. Since the inception of this pandemic, many viral and human protein targets have been identified as potential drug targets. The possible therapeutic strategies target different stages of the viral life cycle from blocking the entry of virions into the cell to inhibit viral replication and through modulating the immune system.
The genome of SARS-CoV-2 encodes various structural and non-structural proteins on which different binding sites have been identified and explored as a potential druggable site. In our study, we included eight different targets. Spike protein promotes the entry of the virus by binding with the host ACE2 receptor, followed by cleavage of the virus by host protein furin and TMPRSS2. 3CLpro, RNA-dependent RNA polymerase (RdRp), Papain-like protease (PLpro), helicase, and endoribonuclease help in the viral replication and progression of the life cycle. 3CLpro also known as main protease or NSP5 is an enzyme that cleaves 11 different sites on pp1a and pp1ab for further cleavage of non-structural proteins Nsp4-nsp16. This non-structural protein includes RdRp, helicase, endoribonuclease, exonuclease, and 2′-O-methyltransferase which are important for the viral genome [39]. RdRp (Nsp12) plays important role in the replication and transcription process of the virus [40]. PLpro is another important protein that cleaves the N-terminal of pp1 and pp1b to generate Nsp1 to Nsp3 with help of 3CLpro. PLpro is also known for modulating the innate immunity of host cells. Helicase is involved in the uncoiling of double-stranded oligonucleotides in an NTP-dependent manner in the 5′-3′ direction and also has a metal-binding domain at N-terminal. Endoribonuclease plays important role in shrinking of host defence mechanism so inhibition of this target may increase the threshold of host immune response [41].
The natural products offer a variety of bioactive substances of different classes and some of the classes such as flavonoids, alkaloids, and peptides have been already tested either in-silico or invitro. Natural products have shown promising results against various other viral diseases [42]. Besides, natural products are safer in terms of toxicity profile as compared to synthetic compounds. Considering these facts, natural products can provide an innovative solution to fight against SARS-CoV-2. Theaflavins are the most abundant constituents found in black tea. Theaflavins are known to have a wide range of pharmacological actions including their anti-viral effect. The activity of theaflavins has been reported against different viral species [43] which includes influenza A & B, Calici, Sindbis, TMV, HSV, rota, corona, HCV, and HIV-1. In a study of HSV-1 virus infection, theaflavins had shown strong inhibition of the viral life cycle, and TF3 was found more potent as compared to TF1 and TF2[15]. In another study against the influenza virus, TF3 was found to be better than the other theaflavin derivatives [44]. Promising results of anti-viral activities of TF3 in the past had encouraged to explore its role through in-silico methods on various targets of SARS-CoV-2 that plays an important role in the viral life cycle. Molecular docking studies are a very useful technique to predict binding affinity and molecular mechanisms. The docking scores of TF3 were found promising with all the receptors that were included in the study. Except with furin, the docking scores of the TF3 were higher than positive controls (Table 2). The binding free energy (MM-GBSA) of all the TF3-protein complexes was also found higher (Table 2). The reason behind good binding affinity as well as binding free energy is presence of polyphenolic as well keto groups. Each group provides favorable environment for H-Bond interaction that helps in strong binding with receptor. Molecular dynamic study results showed that the interactions with CLpro, RdRp, endoribonuclease, and spike protein were more stable than their positive controls for a longer duration than with other receptors. In-silico analysis on the pharmacokinetic and safety parameters of TF3 was very encouraging (Table 4).
The inclusion of positive controls on various targets as a comparator of TF3 through molecular docking and dynamics studies gives a more precise evaluation of TF3 in terms of binding pattern. The active site residues selection for all our targets was either based on the binding site of the cognate ligand in the protein structure or the interacting residues of SARS-CoV-2 with the host protein and residues interacting within an enzyme complex.
5 Conclusion
The world has witnessed a never seen before pandemic since December 2019 and still, the search for a treatment for SARS-CoV-2 is going on. Various drugs have already been tried but none of them proved to be completely curative. Theaflavin-3,3′-digallate is present in commonly used beverage, black tea. Through this study, we tried to evaluate the role of theaflavin-3,3′-digallate on multiple targets of SARS-CoV-2. The docking scores of TF3 were found promising with all the receptors that were included in the study except furin. The binding free energy (MM-GBSA) of all the TF3-protein complexes was also found higher. Molecular dynamic study results showed that the interactions with CLpro, RdRp, endoribonuclease, and spike protein were more stable than their positive controls for a longer duration than with other receptors. In-silico analysis on the pharmacokinetic and safety parameters of TF3 was very encouraging. The positive in-silico results which were obtained on various pharmacodynamic and pharmacokinetic parameters, give a ray of hope as a potential therapeutic drug to this rapidly spreading disease. Hence, we strongly recommend the further exploration of this compound through in-vitro and in-vivo studies.
Availability of Data and Material
Supplementary data is available.
Abbreviations
- ACE2:
-
Angiotensin-converting enzyme 2
- ADMET:
-
Absorption, distribution, metabolism, excretion, toxicity
- CLpro:
-
Chymotrypsin like protease
- Cog:
-
Cognate
- PLpro:
-
Papain like protease
- RBD:
-
Receptor binding domain
- RdRp:
-
RNA dependant RNA polymerase
- Rem:
-
Remdesivir
- TF3:
-
Theaflavin-3,3′-digallate
- TMPRSS2:
-
Transmembrane protease, serine 2
References
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2012-7
Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC (2020) Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285. https://doi.org/10.1038/s41586-020-2169-0
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, NY) 367(6485):1444–1448. https://doi.org/10.1126/science.abb2762
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, NY) 367(6483):1260–1263. https://doi.org/10.1126/science.abb2507
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. https://doi.org/10.1038/s41586-020-2180-5
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281-292.e286. https://doi.org/10.1016/j.cell.2020.02.058
Giordano D, De Masi L, Argenio MA, Facchiano A (2021) Structural dissection of viral spike-protein binding of SARS-CoV-2 and SARS-CoV-1 to the human angiotensin-converting enzyme 2 (ACE2) as cellular receptor. Biomedicines 9(8):1038
Shang W, Yang Y, Rao Y, Rao X (2020) The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. Npj Vaccines 5(1):18. https://doi.org/10.1038/s41541-020-0170-0
Vkovski P, Kratzel A, Steiner S, Stalder H, Thiel V (2021) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19(3):155–170. https://doi.org/10.1038/s41579-020-00468-6
Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C (2008) SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res 18(2):290–301. https://doi.org/10.1038/cr.2008.15
Prasansuklab A, Theerasri A, Rangsinth P, Sillapachaiyaporn C, Chuchawankul S, Tencomnao T (2020) Anti-COVID-19 drug candidates: a review on potential biological activities of natural products in the management of new coronavirus infection. J Tradit Complement Med. https://doi.org/10.1016/j.jtcme.2020.12.001
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23(1):3–20. https://doi.org/10.1038/s41580-021-00418-x
Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C (2019) From SARS to MERS, thrusting coronaviruses into the Spotlight. Viruses. https://doi.org/10.3390/v11010059
Lu W, Zheng BJ, Xu K, Schwarz W, Du L, Wong CK, Chen J, Duan S, Deubel V, Sun B (2006) Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci USA 103(33):12540–12545. https://doi.org/10.1073/pnas.0605402103
Wong NA, Saier MH Jr (2021) The SARS-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int J Mol Sci. https://doi.org/10.3390/ijms22031308
South AM, Brady TM, Flynn JT (2020) ACE2 (angiotensin-converting enzyme 2), COVID-19, and ACE inhibitor and ang II (angiotensin II) receptor blocker use during the pandemic: the pediatric perspective. Hypertension 76(1):16–22. https://doi.org/10.1161/hypertensionaha.120.15291
Alipoor SD, Mortaz E, Jamaati H, Tabarsi P, Bayram H, Varahram M, Adcock IM (2021) COVID-19: molecular and cellular response. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2021.563085
Higdon JV, Frei B (2003) Tea catechins and polyphenols: health effects, metabolism, and antioxidant functions. Crit Rev Food Sci Nutr 43(1):89–143. https://doi.org/10.1080/10408690390826464
Lambert JD, Yang CS (2003) Mechanisms of cancer prevention by tea constituents. J Nutr 133(10):3262s–3267s. https://doi.org/10.1093/jn/133.10.3262S
de Oliveira A, Prince D, Lo CY, Lee LH, Chu TC (2015) Antiviral activity of theaflavin digallate against herpes simplex virus type 1. Antiviral Res 118:56–67. https://doi.org/10.1016/j.antiviral.2015.03.009
Escalante DE, Ferguson DM (2021) Structural modeling and analysis of the SARS-CoV-2 cell entry inhibitor camostat bound to the trypsin-like protease TMPRSS2. Med Chem Res. https://doi.org/10.1007/s00044-021-02708-7
Greenwood JR, Calkins D, Sullivan AP, Shelley JC (2010) Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des 24(6–7):591–604. https://doi.org/10.1007/s10822-010-9349-1
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12(1):281–296. https://doi.org/10.1021/acs.jctc.5b00864
Hussain M, Jabeen N, Amanullah A, Baig AA, Aziz B, Shabbir S, Raza F, Uddin N (2020) Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS Microbiol 6(3):350–360. https://doi.org/10.3934/microbiol.2020021
Heo L, Park H, Seok C (2013) GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41(Web Server issue):W384-388. https://doi.org/10.1093/nar/gkt458
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1):12–21. https://doi.org/10.1107/s0907444909042073
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234. https://doi.org/10.1007/s10822-013-9644-8
Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55(2):351–367. https://doi.org/10.1002/prot.10613
Yan L, Zhang Y, Ge J, Zheng L, Gao Y, Wang T, Jia Z, Wang H, Huang Y, Li M, Wang Q, Rao Z, Lou Z (2020) Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun 11(1):5874. https://doi.org/10.1038/s41467-020-19770-1
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21):6177–6196. https://doi.org/10.1021/jm051256o
Rastelli G, Del Rio A, Degliesposti G, Sgobba M (2010) Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31(4):797–810. https://doi.org/10.1002/jcc.21372
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935. https://doi.org/10.1063/1.445869
Davidchack RL, Handel R, Tretyakov MV (2009) Langevin thermostat for rigid body dynamics. J Chem Phys 130(23):234101. https://doi.org/10.1063/1.3149788
Plewczynski D, Łaźniewski M, Augustyniak R, Ginalski K (2011) Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J Comput Chem 32(4):742–755. https://doi.org/10.1002/jcc.21643
Thompson DC, Humblet C, Joseph-McCarthy D (2008) Investigation of MM-PBSA rescoring of docking poses. J Chem Inf Model 48(5):1081–1091. https://doi.org/10.1021/ci700470c
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem 32(5):866–877. https://doi.org/10.1002/jcc.21666
Kuhn B, Gerber P, Schulz-Gasch T, Stahl M (2005) Validation and use of the MM-PBSA approach for drug discovery. J Med Chem 48(12):4040–4048. https://doi.org/10.1021/jm049081q
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733. https://doi.org/10.1056/NEJMoa2001017
Hegyi A, Friebe A, Gorbalenya AE, Ziebuhr J (2002) Mutational analysis of the active centre of coronavirus 3C-like proteases. J Gen Virol 83(Pt 3):581–593. https://doi.org/10.1099/0022-1317-83-3-581
Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP, Ferron F, Gorbalenya AE, Canard B (2006) A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J 25(20):4933–4942. https://doi.org/10.1038/sj.emboj.7601368
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 10(5):766–788. https://doi.org/10.1016/j.apsb.2020.02.008
Antonio AdS, Wiedemann LSM, Veiga-Junior VF (2020) Natural products’ role against COVID-19. RSC Adv 10(39):23379–23393. https://doi.org/10.1039/D0RA03774E
Mhatre S, Srivastava T, Naik S, Patravale V (2020) Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomedicine. https://doi.org/10.1016/j.phymed.2020.153286
Zu M, Yang F, Zhou W, Liu A, Du G, Zheng L (2012) In vitro anti-influenza virus and anti-inflammatory activities of theaflavin derivatives. Antiviral Res 94(3):217–224. https://doi.org/10.1016/j.antiviral.2012.04.001
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
conceptualization: MJ and AS. Data handling, analysis and presentation: MJ, AA and AS. Experimental design: MJ, AA and AS. Study material and equipment: MJ. Study validation, supervision, draft preparation, study consultation, writing and reviewing and project administration: MJ, AA and AS.
Corresponding author
Ethics declarations
Conflicts of interest
All the authors declared that they have no conflict of interest and no competing interest.
Ethical statement.
Not applicable.
Ethics approval
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jain, M., Anand, A. & Shah, A. Exploring the Potential Role of Theaflavin-3,3′-Digallate in Inhibiting Various Stages of SARS-CoV-2 Life Cycle: An In-Silico Approach. Chemistry Africa 5, 883–898 (2022). https://doi.org/10.1007/s42250-022-00376-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-022-00376-7