Abstract
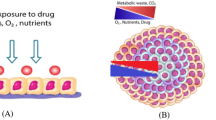
The conventional microwell-based platform for construction of organoid models exhibits limitations in precision oncology applications because of low-speed growth and high variability. Here, we established organoid models on a nested array chip for fast and reproducible drug testing using 50% matrigel. First, we constructed mouse small intestinal and colonic organoid models. Compared with the conventional microwell-based platform, the mouse organoids on the chip showed accelerated growth and improved reproducibility due to the nested design of the chip. The design of the chip provides miniaturized and uniform shaping of the matrigel that allows the organoid to grow in a concentrated and controlled manner. Next, a patient-derived organoid (PDO) model from colorectal cancer tissues was successfully generated and characterized on the chip. Finally, the PDO models on the chip, from three patients, were implemented for high-throughput drug screening using nine treatment regimens. The drug sensitivity testing on the PDO models showed good quality control with a coefficient of variation under 10% and a Z' factor of more than 0.7. More importantly, the drug responses on the chip recapitulate the heterogeneous response of individual patients, as well as showing a potential correlation with clinical outcomes. Therefore, the organoid model coupled with the nested array chip platform provides a fast and reproducible means for predicting drug responses to accelerate precise oncology.
Graphic abstract






Similar content being viewed by others
References
Chae YK, Pan AP, Davis AA et al (2017) Path toward precision oncology: review of targeted therapy studies and tools to aid in defining “actionability” of a molecular lesion and patient management support. Mol Cancer Ther 16(12):2645–2655. https://doi.org/10.1158/1535-7163.Mct-17-0597
Tran A, Klossner Q, Crain T et al (2020) Shifting, overlapping and expanding use of “precision oncology” terminology: a retrospective literature analysis. BMJ Open 10(6):e036357. https://doi.org/10.1136/bmjopen-2019-036357
Mosele F, Remon J, Mateo J et al (2020) Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO precision medicine working group. Ann Oncol 31(11):1491–1505. https://doi.org/10.1016/j.annonc.2020.07.014
Gulilat M, Lamb T, Teft WA et al (2019) Targeted next generation sequencing as a tool for precision medicine. BMC Med Genomics 12:81. https://doi.org/10.1186/s12920-019-0527-2
Asada K, Kaneko S, Takasawa K et al (2021) Integrated analysis of whole genome and epigenome data using machine learning technology: toward the establishment of precision oncology. Front Oncol 11:666937. https://doi.org/10.3389/fonc.2021.666937
Souza GR, Molina JR, Raphael RM et al (2010) Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol 5(4):291–296. https://doi.org/10.1038/nnano.2010.23
Boughey JC, Suman VJ, Yu J et al (2021) Patient-derived xenograft engraftment and breast cancer outcomes in a prospective neoadjuvant study (beauty). Clin Cancer Res 27(17):4696–4699. https://doi.org/10.1158/1078-0432.Ccr-21-0641
Li M, Belmonte JCI (2019) Organoids—preclinical models of human disease. New Engl J Med 380(6):569–579. https://doi.org/10.1056/NEJMra1806175
Method of the year 2017: organoids. Nat Methods 15(1):1. https://doi.org/10.1038/nmeth.4575
Koh V, Chakrabarti J, Torvund M et al (2021) Hedgehog transcriptional effector GLI mediates mTOR-induced PD-L1 expression in gastric cancer organoids. Cancer Lett 518:59–71. https://doi.org/10.1016/j.canlet.2021.06.007
Bose S, Clevers H, Shen X (2021) Promises and challenges of organoid-guided precision medicine. Med 2(9):1011–1026. https://doi.org/10.1016/j.medj.2021.08.005
Borries M, Barooji YF, Yennek S et al (2020) Quantification of visco-elastic properties of a matrigel for organoid development as a function of polymer concentration. Front Phys 8:579168. https://doi.org/10.3389/fphy.2020.579168
Gao D, Vela I, Sboner A et al (2014) Organoid cultures derived from patients with advanced prostate cancer. Cell 159(1):176–187. https://doi.org/10.1016/j.cell.2014.08.016
Vlachogiannis G, Hedayat S, Vatsiou A et al (2018) Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359(6378):920–926. https://doi.org/10.1126/science.aao2774
Boehnke K, Iversen PW, Schumacher D et al (2016) Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J Biomol Screen 21(9):931–941. https://doi.org/10.1177/1087057116650965
Driehuis E, Kretzschmar K, Clevers H (2020) Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc 15(10):3380–3409. https://doi.org/10.1038/s41596-020-0379-4
Geuens T, van Blitterswijk CA, LaPointe VLS (2020) Overcoming kidney organoid challenges for regenerative medicine. NPJ Regen Med 5(1):8. https://doi.org/10.1038/s41536-020-0093-4
Brandenberg N, Hoehnel S, Kuttler F et al (2020) High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat Biomed Eng 4(9):863–874. https://doi.org/10.1038/s41551-020-0565-2
Hou Y, Ai XN, Zhao L et al (2020) An integrated biomimetic array chip for high-throughput co-culture of liver and tumor microtissues for advanced anticancer bioactivity screening. Lab Chip 20(14):2482–2494. https://doi.org/10.1039/d0lc00288g
Xiao RR, Lv T, Tu X et al (2021) An integrated biomimetic array chip for establishment of collagen-based 3D primary human hepatocyte model for prediction of clinical drug-induced liver injury. Biotechnol Bioeng 118(12):4687–4698. https://doi.org/10.1002/bit.27931
Park SE, Georgescu A, Huh D (2019) Organoids-on-a-chip. Science 364(6444):960–965. https://doi.org/10.1126/science.aaw7894
Wan L, Neumann CA, Leduc PR (2020) Tumor-on-a-chip for integrating a 3D tumor microenvironment: chemical and mechanical factors. Lab Chip 20(5):873–888. https://doi.org/10.1039/c9lc00550a
Wasson EM, Dubbin K, Moya ML (2021) Go with the flow: modeling unique biological flows in engineered in vitro platforms. Lab Chip 21(11):2095–2120. https://doi.org/10.1039/d1lc00014d
Ai XN, Zhao L, Lu YY et al (2020) Integrated array chip for high-throughput screening of species differences in metabolism. Anal Chem 92(17):11696–11704. https://doi.org/10.1021/acs.analchem.0c01590
Ai XN, Lu WB, Zeng KW et al (2018) Microfluidic coculture device for monitoring of inflammation induced myocardial injury dynamics. Anal Chem 90(7):4485–4494. https://doi.org/10.1021/acs.analchern.7b04833
Hu YW, Sui XZ, Song F et al (2021) Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat Commun 12(1):2581. https://doi.org/10.1038/s41467-021-22676-1
Jiang SW, Zhao HR, Zhang WJ et al (2020) An automated organoid platform with inter-organoid homogeneity and inter-patient heterogeneity. Cell Rep Med 1(9):100161. https://doi.org/10.1016/j.xcrm.2020.100161
Rawal P, Tripathi DM, Ramakrishna S et al (2021) Prospects for 3D bioprinting of organoids. Bio-Des Manuf 4(3):627–640. https://doi.org/10.1007/s42242-020-00124-1
Van Zundert I, Fortuni B, Rocha S (2020) From 2D to 3D cancer cell models—the enigmas of drug delivery research. Nanomaterials 10(11):2236. https://doi.org/10.3390/nano10112236
Wilson SS, Mayo M, Melim T et al (2021) Optimized culture conditions for improved growth and functional differentiation of mouse and human colon organoids. Front Immunol 11:547102. https://doi.org/10.3389/fimmu.2020.547102
Grabinger T, Luks L, Kostadinova F et al (2014) Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis 5(5):e1228. https://doi.org/10.1038/cddis.2014.183
Agarwal T, Celikkin N, Costantini M et al (2021) Recent advances in chemically defined and tunable hydrogel platforms for organoid culture. Bio-Des Manuf 4(3):641–674. https://doi.org/10.1007/s42242-021-00126-7
Choi JI, Jang SI, Hong J et al (2021) Cancer-initiating cells in human pancreatic cancer organoids are maintained by interactions with endothelial cells. Cancer Lett 498:42–53. https://doi.org/10.1016/j.canlet.2020.10.012
Takigawa H, Kitadai Y, Shinagawa K et al (2016) Multikinase inhibitor regorafenib inhibits the growth and metastasis of colon cancer with abundant stroma. Cancer Sci 107(5):601–608. https://doi.org/10.1111/cas.12907
Backes Y, Seerden TCJ, Van Gestel RSFE et al (2019) Tumor seeding during colonoscopy as a possible cause for metachronous colorectal cancer. Gastroenterology 157(5):1222–1232. https://doi.org/10.1053/j.gastro.2019.07.062
Heydari Z, Moeinvaziri F, Agarwal T et al (2021) Organoids: a novel modality in disease modeling. Bio-Des Manuf 4(4):689–716. https://doi.org/10.1007/s42242-021-00150-7
Lau HCH, Kranenburg O, Xiao HP et al (2020) Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastro Hepat 17(4):203–222. https://doi.org/10.1038/s41575-019-0255-2
Skala MC, Deming DA, Kratz JD (2022) Technologies to assess drug response and heterogeneity in patient-derived cancer organoids. Annu Rev Biomed Eng 24:157–177. https://doi.org/10.1146/annurev-bioeng-110220-123503
Sato T, Vries RG, Snippert HJ et al (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244):262–265. https://doi.org/10.1038/nature07935
Kozlowski MT, Crook CJ, Ku HT (2021) Towards organoid culture without matrigel. Commun Biol 4(1):1387. https://doi.org/10.1038/s42003-021-02910-8
Jee JH, Lee DH, Ko J et al (2019) Development of collagen-based 3D matrix for gastrointestinal tract-derived organoid culture. Stem Cells Int 2019:8472712. https://doi.org/10.1155/2019/8472712
Sun Q, Zhou J, Zhang Z et al (2014) Discovery of fruquintinib, a potent and highly selective small molecule inhibitor of VEGFR 1, 2, 3 tyrosine kinases for cancer therapy. Cancer Biol Ther 15(12):1635–1645. https://doi.org/10.4161/15384047.2014.964087
Ding S, Hsu C, Wang Z et al (2022) Patient-derived micro-organospheres enable clinical precision oncology. Cell Stem Cell 29(6):905–917. https://doi.org/10.1016/j.stem.2022.04.006
Bose S, Clevers H, Shen XL (2021) Promises and challenges of organoid-guided precision medicine. Med-Cambridge 2(9):1011–1026. https://doi.org/10.1016/j.medj.2021.08.005
Van De Wetering M, Francies HE, Francis JM et al (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161(4):933–945. https://doi.org/10.1016/j.cell.2015.03.053
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 82174086), the Beijing Natural Science Foundation (No. 7222273), the Beijing Xisike Clinical Oncology Research Foundation (Nos. Y-xsk2021-0004 and Y-XD202001-0172), the Youth Talents Promotion Project of China Association of Chinese Medicine (No. 2020-QNRC2-08), the Clinical Medicine Plus X-Young Scholars Project of Peking University (No. BMU2021MX009), and the Peking University People’s Hospital Research and Development Funds (No. RDY2020-18).
Author information
Authors and Affiliations
Contributions
XA, YC, and RX contributed to conceptualization; XA, YC, RX, JL, YL, and GX provided methodology; XA, YC, RX, YZ, XY, ZS, BL, and KS provided resources; XA, YC, RX, and YW carried out data analysis; XA, YC, and YW performed writing—original draft preparation; XA performed writing—review and editing; XA and YY performed supervision; YC, XA, and YY contributed to funding acquisition. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
XA is the scientific advisor at Beijing Daxiang Biotech. RX, JL and YW are current employees at Beijing Daxiang Biotech. YL and GX are current employees in Merck Innovation Hub (Guangdong) Co., Ltd..
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethical committees of Peking University People’s Hospital (Ethics approval number: 2021PHB148-001) and registered in ClinicalTrial.gov (NCT04996355). Informed consent was obtained from all patients for being included in the study.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cui, Y., Xiao, R., Zhou, Y. et al. Establishment of organoid models based on a nested array chip for fast and reproducible drug testing in colorectal cancer therapy. Bio-des. Manuf. 5, 674–686 (2022). https://doi.org/10.1007/s42242-022-00206-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42242-022-00206-2