Abstract
A root disease in plantations of Pinus radiata and Pinus pinaster, where trees died in distinct patches, was present in the Western Cape province of South Africa during the 1970s and 1980s. Phytophthora cinnamomi was initially believed to be the cause, but the disease was later ascribed to the insect-associated fungus Leptographium serpens, a fungal species residing in the Ophiostomatales. Doubt regarding the cause of the disease was raised in a later study due to the fact that most Leptographium spp., particularly those that colonise ray parenchyma tissues, which is the case for L. serpens, are not typically primary disease agents. In this study, cultures of an unidentified sterile fungus collected from the dying trees were revived and identified using DNA sequencing methods, which were not available when the disease was first studied. These cultures were identified as the pyrophillic pathogen Rhizina undulata, well-known to cause patch death of conifers in South Africa and elsewhere in the world. While the patches of dying trees no longer exist and the disease cannot be newly studied, it is most likely that the tree death originally thought to be caused by L. serpens was due primarily to R. undulata. The study provides a vivid example of the value of preserving cultures of fungi for later study and the power of modern techniques to identify fungal pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
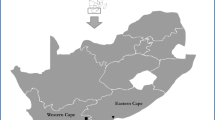
During the 1970s and 1980s, patches of Pinus radiata and Pinus pinaster growing in the Grabouw area of South Africa’s Western Cape province were observed to be diseased (Fig. 1). The disease was typified by trees at the peripheries of these patches gradually dying, while naturally regenerating plants within the dead areas wilted and died during the first few years of growth. At the time when this disease was first observed, the Oomycete root pathogen Phytophthora cinnamomi was recognized as important to agriculture and particularly the Protea (Proteaceae) cut-flower industry of the Western Cape province (Brits et al. 1983). An interest in P. cinnamomi also led to its discovery in a Grabouw forestry nursery (Donald and Broembsen 1977). And in turn, the patches of diseased trees in local plantations were assumed to be due to that pathogen (Wilson 1977).
(A-C) Dying Pinus pinaster trees in the Grabouw plantation, Western Cape Province, South Africa. (B) Infected trees in distinct patches with newly dying trees at the periphery. (C) Naturally regenerated trees within a disease centre dying, as indicated by white arrows. Note: these photographs were taken between 1976 and 1979 in a planation area that no longer exists. (D) Ascocarp of Rhizina undulata commonly found in South African Pinus plantations after fire. These were not found during the disease outbreak in the 1970s and 1980s. (E) Asci and ascospores of R. undulata from a South African collection of the pathogen. Scale bar: E = 100 μm
The dying trees found in the disease centres showed evidence of root disease with actively developing lesions in the cambium of the roots and root collars. Infected wood associated with cambial infection had a distinct grey to black colour that did not penetrate into the wood. No fungal fruiting bodies were found on the ground associated with the dying trees. A fungus was consistently isolated from the leading edges of the actively developing lesions on the diseased roots. It was determined that this fungus was a new species in the genus Verticicladiella and the name Verticicladiella alacris was provided for it (Wingfield and Marasas 1980). Pathogenicity tests with the fungus showed that it gave rise to lesions similar to those found on dying trees, and the disease was subsequently attributed to this fungus (Wingfield and Knox-Davies 1980).
The description of V. alacris relied on morphological characteristics (Wingfield and Marasas 1980). This also led to the fungus being reduced to synonymy with Verticicladiella serpens (Wingfield and Marasas 1981), a fungus associated with diseased roots of Pinus pinea in Italy (Lorenzini and Gambogi 1976). Later, these fungi and other species of Verticicladiella were transferred to the genus Leptographium, a well-represented asexual genus of the Ophiostomatales (Seifert et al. 2013; De Beer et al. 2022). The fungus originally associated with dying Pinus spp. in South Africa was relegated to the name Leptographium serpens (Wingfield 1985). Duong et al. (2012) conducted a detailed study of Leptographium spp. associated with root infections of Pinus spp. in which DNA sequence data for five gene regions were applied. That study showed that the fungus associated with the root disease of the Pinus spp. in the Western Cape was different to L. serpens and the name of the South African fungus was most appropriately Leptographium alacre (Morelet 1985; Duong et al. 2012; De Beer et al. 2022).
There has been a long-standing debate regarding the role that Leptographium spp. can play as the primary causal agents of conifer root disease (Harrington 1988; Wingfield et al. 1988; Jacobs and Wingfield 2001). Three Leptographium spp. have been associated with such diseases including the three varieties of Leptographium wageneri (Wagener and Mielke 1961), Leptographium procerum (Dochinger 1967) and, as mentioned above, L. serpens (Lorenzini and Gambogi 1976). Of these fungi, L. wageneri, the causal agent of black stain root disease is clearly different to the others. This difference lies in the fact that it colonises only the tracheids and does not invade the ray parenchyma tissues (Diamandis et al. 1997; Harrington and Cobb Jr 1983; Wingfield 1986). In this regard, it might be considered a true vascular wilt pathogen. It is thus very different to other species of Leptographium that colonise the ray parenchyma tissues, giving rise to pie-shaped lesions similar to the so-called blue-stain fungi (Jacobs and Wingfield 2001). Such pie-shaped discoloration was not seen in diseased trees from the 1970s and 1980s outbreak, because the activiely infected tissue was in the fresh cambium and would not have developed at this early state of infection. The role of these other Leptographium spp. in root disease is best considered as contributing to tree death rather than being primary disease agents (Jacobs and Wingfield 2001). This then calls to question the role that L. serpens played in causing the root disease of Pinus spp. in the Western Cape province of South Africa in the 1970s and 1980s.
Wingfield et al. (1988) raised the issue that the root disease of P. radiata and P. pinaster in South Africa had not been fully elucidated. In that review, it was mentioned that a sterile fungus, tentatively referred to as a species of Rhizoctonia, had been isolated from the roots of the trees in question, including those from which L. alacre had been isolated. The present study considered the identification of cultures of the sterile fungus isolated in 1984 and referred to by Wingfield et al. (1988), which have been preserved in a culture collection for approximately four decades.
Materials and methods
Cultures
This study relied on the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) housed at the University of Pretoria, Pretoria, South Africa. Four cultures were deposited in the collection in March 1987 by the first author (MJW) of the present paper and were isolated from the roots of dying trees at the periphery of the disease centres and from the roots of the young naturally regenerating trees within the patches. Two of these cultures (CMW 435 and CMW 436) were isolated from Pinus pinaster on September 11, 1984 and two (CMW 413 and CMW 412) were isolated from P. radiata on July 17 and August 8, 1984, respectively. These four cultures were accessed from the collection in August 2020 and plated on 2% malt extract media (20 g Biolab malt extract, 20 g Difco agar, 1 L deionized water). They were incubated in the dark at room temperature and two cultures (CMW 412 and CMW 436) could be revived.
PCR, DNA sequencing, and phylogenetic analyses
DNA was isolated from 7-day-old cultures of isolates CMW 412 and CMW 436 using the Prepman Ultra Sample Preparation Reagent (Thermo Fisher Scientific). The internal transcribed spacer regions 1 and 2 (ITS), including the 5.8 S rRNA region, were amplified using primers ITS1F and ITS4 (Gardes and Bruns 1993; White et al. 1990). PCR mixtures were prepared following the protocols described by Pham et al. (2019). The thermal cycling included an initial denaturation at 95 °C for 5 min followed by 10 primary amplification cycles of 30 s at 95 °C, 30 s at 56 °C, and 60 s at 72 °C, then 30 additional cycles of the same reaction sequence, with a 5 s increase in the annealing step per cycle, and the reactions were completed with a final extension at 72 °C for 10 min. Amplified fragments were treated with ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific). The purified products were sequenced in both directions using the BigDye terminator sequencing kit v. 3.1 (Thermo Fisher Scientific) on an ABI Prism 3100 DNA sequencer (Thermo Fisher Scientific) at the Sequencing Facility of the Faculty of Natural and Agricultural Sciences, University of Pretoria. Geneious Prime v. 2022.1.1 (https://www.geneious.com) was used to assemble and trim the raw sequences, which were deposited in GenBank (Table 1).
Preliminary identification was made by performing a nucleotide BLAST search using the ITS sequences against the NCBI GenBank database (http://www.ncbi.nlm.nih.gov) to identify the isolates to the genus level. Sequences of species closely related to those emerging from this study were sourced from the GenBank database (Table 1). All sequences were aligned with MAFFT v. 7 (Katoh and Standley 2013) and inspected manually using MEGA v. 7 (Kumar et al. 2016). A Maximum-Likelihood (ML) analysis was conducted using RaxML v. 8.2.4 (Stamatakis 2014) on the CIPRES Science Gateway 3.3 (Miller et al. 2011), with default GTR substitution model and 1000 rapid bootstraps. Sequences for Helvella bachu (HKAS 88105) and Helvella calycina (O-253255) were used as outgroups.
Results
PCR, DNA sequencing, and phylogenetic analyses
For both of the South African isolates considered in this study, amplicons of approximately 500 bp were generated for the ITS region. The dataset used in the phylogenetic analyses included 15 ingroup taxa and contained 801 characters, including alignment gaps. When comparing sequences for the isolates considered in this study with those for species in Rhizinaceae, the two South African isolates had identical sequences and formed a well-supported (95%) monophyletic clade in the ML tree (Fig. 2) with all the reference isolates of Rhizina undulata. The isolates were thus identified as that fungus.
Phylogenetic tree based on a Maximum Likelihood (ML) analysis of ITS sequences representing species of Rhizinaceae. The two isolates collected in South Africa in 1984 and deposited in the culture collection (CMW) of FABI are presented in bold face. The countries of origin of the isolates are indicated in red text. Bootstrap values for the ML analyses are indicated at the nodes. Helvella bachu (HKAS 88105) and Helvella calycina (O-253255) represent the outgroup taxa
Discussion
An unknown sterile fungus, isolated from the roots of diseased Pinus spp. in South Africa and thought to be dying due to infection by the Ophiostomatoid fungus L. alacre, was identified in the present study as the root pathogen R. undulata. This fungus is well-known to cause patch death of Pinus spp. as well as other conifers in various parts of the world (Peace 1962; Gibson 1979). It is also well-known in South Africa (Fig. 1), where it can result in a failure to establish new plantations after fire (Gibson 1979; Lundquist 1984). The previous uncertainty (Wingfield et al., 1988) regarding the role of L. alacre in causing the patch death of P. radiata and P. pinaster is thus likely resolved by the results of this study. This arises from the fact that we have identified a well-known root pathogen, typically sterile in culture, that was isolated from actively emerging lesions on the roots of the dying trees, some 40 years after the disease problem was first reported.
An important question arising from this study is what role L. alacre might have played in the root disease as reported by Wingfield and Marasas (1980). It is not possible to consider this question experimentally due to the fact that the plantations where the disease occurred no longer exist. However, a plausible explanation lies in the biology of L. alacre and the manner in which it spreads. This fungus is well-known to be vectored by the root-feeding scolytine (Coleoptera: Curculionidae: Scolytinae) beetle Hylastes angustatus in South Africa (Wingfield and Knox-Davies 1980; Wingfield and Marasas 1980; Zhou et al. 2001). These beetles undergo maturation feeding on the roots of living Pinus trees before they colonise stressed or dying trees. During this feeding period, they transmit their fungal associates, including L. alacre, to the roots. The inoculation trials, conducted in the study of Wingfield and Knox-Davies 1980, showed that L. alacre was able to induce lesions on P. pinaster and P. radiata and that it had relatively high levels of aggressiveness. It is thus likely that L. alacre would have been introduced into these roots due to beetle maturation feeding, outcompeting other fungi including R. undulata, which we now believe was the primary pathogen.
R. undulata is a pyrophillic fungus, requiring heat shock for ascospore germination and for the fungus to become an active pathogen (Jalaluddin 1967). It was first discovered in Europe in the late 1800s and was thought to be the causal agent of la maladie due ronde in France and Ringseuche in Germany (Gremmen 1971). It has commonly been referred to as the “coffee fire fungus” because conifer trees would die in patches activated by the heat of camp fires made to prepare tea or coffee on forestry trails (Peace 1962; Gremmen 1971; Gibson 1979). Thus, an important question arising from the present study is what form of heat stimulus would have activated the pathogen and initiated P. radiata and P. pinaster patch death, the disease previously attributed to L. serpens.
Forms of heat-shock other than forest fires, such as hot coals being thrown from steam locomotives, hot asphalt being laid for the construction of new roads, and accidental fires have been recorded to result in active R. undulata disease centers (Gremmen 1971). Damage due to R. undulata in South Africa is mostly associated with burning of forest waste after clear-felling or accidental fires in plantations of Pinus spp. (Lundquist 1984; Wingfield and Swart 1994). There was no evidence of fires having occurred in any of the patches of dying trees in the Western Cape pine plantations, which would have otherwise provided an early explanation for the problem. While R. undulata typically requires a heat shock to be activated, there is limited evidence to suggest that it can become active in highly acidic soils and in the presence of living conifer roots (Jalaludin 1967). Soils in the areas where the disease was present, known as Table Mountain sandstone (Richards et al. 1997), are sandy and acidic. This provides at least anecdotal support for the fact that R. undulata, which is well-known in Western Cape pine plantations, could have infected the roots of the trees in the absence of a heat shock.
An interesting and important manifestation of the root disease attributed to L. serpens (now L. alacre) was the fact that naturally regenerated trees within the disease patches became diseased and wilted up to approximately one year of age (Fig. 1; Wingfield and Knox-Davies 1980; Wingfield et al. 1988). These trees had active lesions in the freshly infected tissues and from which L. serpens was easily isolated. A plausible explanation for the continuous death of the naturally regenerating trees could relate to the biology of H. angustatus (Tribe 1990). Populations of these insects would have built up in the older dying trees at the periphery of the patches. These insects would then be attracted to the roots of the young naturally regenerating trees where they would undergo maturation feeding and together with L. alacre, cause the young trees to die. Whether these young trees within the disease centres had also died due to R. undulata infection remains unclear.
The fact that the cause of tree death in patches was first attributed to P. cinnamomi was incorrectly based on an intense interest in that pathogen at the time. This view would furthermore have been supported by the fact that P. cinnamomi had been found in a nursery providing planting stock for the plantation where the disease occurred (Brits et al. 1983; Donald and Broembsen 1977) and reports of P. radiata dying in other parts of the world (Newhook 1959; Gibson 1979). Attributing the disease to a species of Leptographium was also erroneous and arose from the consistent association of L. serpens with early disease symptoms and the results of pathogenicity tests with the fungus. If the non-sporulating (sterile) cultures isolated from the roots of dying P. pinaster and P. radiata could have been identified at the time, the cause of the disease would likely have been described differently. Importantly, the results of the present study relied on DNA-based sequencing technology that was not available at the time that the disease problem was first observed. This illustrates the importance of such advancing technologies in the fields of forest pathology and tree health.
This study would not have been possible without access to the unidentified cultures that had been preserved for approximately four decades. Furthermore, the results have provided an explanation for an unexplained tree disease problem; one that has confused the literature regarding Leptographium root diseases for many years. Importantly, it highlights the significance of fungal culture collections and our need to preserve cultures for long periods of time, regardless of their perceived importance at their time of collection. There are few formally recognized and financially subsidized culture collections globally and none of these would consider preserving cultures of unnamed fungi such as those that made the present study possible. The question thus arises as to whether novel approaches, including new technologies, should not be considered such that larger and more diverse living fungal cultures could be maintained for future study. Amongst other advantages, this would advance the study of microbes including fungi, beyond a largely comparative taxonomic focus.
Data availability
The sequences that were generated for this study have been deposited at the NCBI. The accession numbers will be made available upon publication.
References
Brits GJ, Jacobs G, Vogts MM (1983) Domestication of fynbos Proteaceae as a floricultural crop. Bothalia 14:641–646
Chitrampalam P, Olsen MW (2014) Genetic diversity and fungicide sensitivity of Phymatotrichopsis omnivora isolates from cotton in Arizona. Research Report
De Beer ZW, Procter M, Wingfield MJ, Marincowitz S, Duong TA (2022) Generic boundaries in the Ophiostomatales, reconsidered and revised. Stud Mycol 101:57–120
Diamandis S, Epstein L, Cobb F Jr, Popenuck T, Hecht-Poinar E (1997) Development of Leptographium wageneri on root surfaces and other substrata. Eur J for Pathol 27:381–390
Dochinger LS (1967) Leptographium root decline of eastern white pine. Phytopathology 57(8):3340
Donald DGM, von Broembsen SL (1977) The control of Phytophthora cinnamomi Rands in a south african forest nursery. South Afr for J 100:50–55
Duong TA, Wingfield BD, Wingfield MJ (2012) Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia 104:715–732
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for Basidiomycetes- application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118
Gibson IAS (1979) Diseases of forest trees widely planted as exotics in the tropics and southern hemisphere. Part II. The genus Pinus. Commonwealth Forestry Institute, Oxford, UK
Gremmen J (1971) Rhizina undulata: a review of research in the Netherlands. Eur J Plant Pathol 1:1–6
Harrington TC (1988) Leptographium species, their distributions, hosts and insect vectors. In: Harrington TC, Cobb FW Jr (eds) Leptographium root diseases on conifers. American Phytopathological Society Press, St Paul USA, pp 1–39
Harrington T, Cobb F Jr (1983) Pathogenicity of Leptographium and Verticicladiella spp. isolated from roots of western north American confiers. Phytopathology 73:596–599
Healy RA, Arnold AE, Bonito G, Huang YL, Lemmond B, Pfister DH, Smith ME (2021) Endophytism and endolichenism in Pezizomycetes: the exception or the rule? New Phytol 233(5):1974–1983
Hughes KW, Matheny PB, Miller AN, Petersen RH, Iturriaga TM, Johnson KD, Methven AS, Raudabaugh DB, Swenie RA, Bruns TD (2020) Pyrophilous fungi detected after wildfires in the Great Smoky Mountains National Park expand known species ranges and biodiversity estimates. Mycologia 112(4):677–698
Jacobs K, Wingfield MJ (2001) Leptographium species: tree pathogens, insect associates and agents of blue stain. American Phytopathological Soicety Press, St Paul, USA, p 207
Jalaluddin M (1967) Studies on Rhizina undulata: I. Mycelial growth and ascospore germination. Trans Br Mycol Soc 50:449–459
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee SK, Lee JK, Kim KH, Lee SK, Lee SY (2007) PCR detection and sequence analysis of the rDNA ITS regions of Rhizina undulata. J Korean for Soc 96(4):425–431
Lorenzini G, Gambogi P (1976) Decline of Pinus pinea associated with the presence of Verticicladiella spp. (preliminary note). Informatore Fitopatologico 5:5–8
Lundquist JE (1984) The occurrence and distribution of Rhizina root rot in South Africa and Swaziland. South Afr for J 131:22–24
Miller MA, Pfeiffer W, Schwartz T (2011) Creating the CIPRES science gateway for inference of large phylogenetic trees In: Proceedings of the 2011 TeraGrid Conference: extreme digital discovery, pp 1–8
Morelet M (1985) Nouveaux binomes chez les Deuteromycotina. Ann de la Société des Sci Nat et d’Archéologie de Toulon et du Var 37(4):233–234
Newhook FJ (1959) The association of Phytophthora spp. with mortality of Pinus radiata and other conifers: I. symptoms and epidemiology in shelterbelts. New Z J Agric Res 2:808–843
Peace TR (1962) Pathology of trees and shrubs, with special reference to Britain. Oxford University Press, UK
Pham NQ, Barnes I, Chen S, Liu F, Dang QN, Pham TQ, Lombard L, Crous PW, Wingfield MJ (2019) Ten new species of Calonectria from Indonesia and Vietnam. Mycologia 111:78–102
Richards MB, Stock WD, Cowling RM (1997) Soil nutrient dynamics and community boundaries in the fynbos vegetation of South Africa. Plant Ecol 130:143–153
Seifert KA (2013) In: De Beer ZW, Wingfield MJ (eds) The ophiostomatoid fungi: expanding frontiers, vol 12. CBS-KNAW Fungal Biodiversity Series
Skrede I, Carlsen T, Schumacher T (2017) A synopsis of the saddle fungi (Helvella: Ascomycota) in Europe-species delimitation, taxonomy and typification. Pers : Mol Phylogeny Evol Fungi 39(1):201–253
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tribe G (1990) Phenology of Pinus radiata log colonization by the pine bark beetle Hylastes angustatus (Herbst) (Coleoptera: Scolytidae) in the south-western Cape Province. J Entomol Soc South Afr 53:93–100
Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 91(1):23–36
Wagener W, Mielke J (1961) A staining-fungus root disease of ponderosa, Jeffrey and pinyon pines. Plant Dis Report 45(11):831–835
White TJ, Bruns T, Lee S, Taylor JL (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA, pp 315–322
Wilson A (1977) A new threat to pine plantations. Forestry News 1/78. The Department of Forestry, Pretoria, South Africa
Wingfield MJ (1985) Reclassification of Verticicladiella based on conidial development. Trans Br Mycol Soc 85:81–93
Wingfield MJ (1986) Pathogenicity of Leptographium procerum and L. terebrantis on Pinus strohus seedlings and established trees. Eur J for Pathol 16:299–308
Wingfield MJ, Knox-Davies PS (1980) Root disease, associated with Verticicladiella alacris, of pines in South Africa. Plant Dis 64:569–571
Wingfield MJ, Marasas WFO (1980) Verticicladiella alacris sp. nov., associated with a root disease of pines in South Africa. Trans Br Mycol Soc 75:21–28
Wingfield MJ, Marasas WFO (1981) Verticicladiella alacris, a synonym of V. serpens. Trans Br Mycol Soc 76:508–510
Wingfield MJ, Swart WJ (1994) Integrated management of forest tree diseases in South Africa. For Ecol Manage 65:11–16
Wingfield MJ, Capretti P, MacKenzie M (1988) Leptographium spp as root pathogens of conifers: an international perspective. In: Harrington TC, Cobb FW Jr (eds) Leptographium root diseases on conifers. American Phytopathological Society Press, St Paul, USA, pp 113–128
Zhao Q, Sulayman M, Zhu XT, Zhao YC, Yang ZL, Hyde KD (2016) Species clarification of the culinary Bachu mushroom in western China. Mycologia 108(4):828–836
Zhou XD, De Beer ZW, Wingfield BD, Wingfield MJ (2001) Ophiostomatoid fungi associated with three pine-infesting bark beetles in South Africa. Sydowia 53:290–300
Acknowledgements
This study benefitted from various groups including the members of the Tree Protection Co-operative Programme (TPCP) and the National Research Foundation (NRF) in South Africa that have provided funding over many years to support the maintenance of cultures in the collection of the Forestry and Agricultural Biotechnology Institute (FABI) at the University of Pretoria. We are also grateful to colleagues that participated in studies on the disease discussed in this paper and who are acknowledged in previous publications.
Funding
This study benefitted from various groups including the members of the Tree Protection Co-operative Programme (TPCP) and the National Research Foundation (NRF) in South Africa that have provided funding over many years to support the maintenance of cultures in the collection of the Forestry and Agricultural Biotechnology Institute (FABI) at the University of Pretoria.
Open access funding provided by University of Pretoria.
Author information
Authors and Affiliations
Contributions
MJW, BDW and AW conceived and designed the study ; NQP and TAD executed the sequencing and phylogenetic analyses and SM maintained cultures. MJW wrote the first draft of the paper and all authors contributed to its completion.
Corresponding author
Ethics declarations
Statements & declarations
The authors have no competing interests to declare that are relevant to the content of this article.
Conflict of interest
The authors have no conflicting interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wingfield, M.J., Pham, N.Q., Marincowitz, S. et al. Blast from the past: a study of decades-old fungal cultures resolves a long-standing tree disease mystery. J Plant Pathol 106, 377–384 (2024). https://doi.org/10.1007/s42161-023-01502-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-023-01502-1