Abstract
In the Limpopo Province of South Africa, the sustainable production of tomatoes is threatened by a soilborne disease known as Verticillium wilt. Limited information is available regarding the Verticillium species involved, their genetic diversity and aggressiveness. Therefore, a pathogen survey was conducted in the three major tomato production regions in Limpopo (six fields in the Lowveld and two fields each in the Highveld and Soutpansberg). Verticillium dahliae was identified as the only Verticillium species associated with Verticillium wilt of tomatoes. Conventional vegetative compatibility group (VCG) testing identified 38 isolates as VCG 4B and two isolates as VCG 2B. Pathotype-genotype specific PCR analysis indicated that all the isolates belonged to the non-defoliating pathotype, specifically genotype C. These characteristics allowed for the classification of almost all the isolates from the Lowveld, Highveld and Soutpansberg regions into clonal lineage 4B (38 isolates). The exception was for two isolates from the Lowveld that belonged to lineage 2B824. Based on a race-specific PCR analysis, all 40 isolates belonged to race 2. Sequencing of the race-specific PCR amplicon revealed the presence of two haplotypes namely I and II. Evaluation of seven inoculation methods (agar plug, millet, node-inoculation, root-dip, sand-bran, soil-drench and toothpick methods) using three isolates showed that the root-dip method was the only method that consistently identified all the isolates as pathogenic based on disease severity as well as an increase in plant height. Assessment of the aggressiveness of the 40 characterised V. dahliae isolates showed that the isolates varied in aggressiveness to the cv. Floradade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
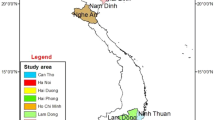
Tomato is one of the most important vegetable crops grown worldwide, and it is the second most important in South Africa. In South Africa, tomatoes are produced in six provinces (Eastern Cape, KwaZulu-Natal, Limpopo, Mpumalanga, North West and Western Cape). The main production region is the Limpopo Province, which is divided into the Soutpansberg, Laeveld and Highveld regions, where 75% (3590 ha) of the country’s tomatoes are grown. Tomato production contributes significantly to the economy of the region and South Africa. A vascular soilborne disease known as Verticillium wilt, which is caused by a few Verticillium spp., threatens the sustainable production of tomatoes in South Africa.
Internationally, ten Verticillium spp. are known to cause vascular wilts on a wide host range including agricultural crops, woody hosts and ornamentals (Inderbitzin et al. 2011). The five species associated with tomato are V. dahliae, V. isaacii, V. nonalfalfae, V. tricorpus and V. zaregamsianum (Inderbitzin and Subbarao 2014). In South Africa, only V. dahliae and V. tricorpus have been reported. Verticillium dahliae is known to occur in all production regions whereas V. tricorpus has only been recorded from the Skeerpoort area in the North West Province (Gorter 1977; Uys and Rong 1995; Uys et al. 1997). The aforementioned South African studies were all conducted before it became known that sequence data are required for accurate species identifications since some species cannot be differentiated morphologically. For example, V. isaacii and V. tricorpus are morphologically indistinguishable. DNA regions that have been utilised for species identification include actin (ACT), elongation factor 1-alpha (EF), glyceraldehyde-3-phosphate dehydrogenase (GPD), tryptophan synthase (TS) and the ribosomal internal transcribed spacer region (ITS) (Inderbitzin et al. 2011). These genomic regions, depending on the species involved, have been used to develop species-specific primers to facilitate rapid and cost-effective identification of the ten known Verticillium species (Inderbitzin et al. 2013).
Verticillium dahliae tomato isolates can be differentiated into two races according to their virulence (gene-for-gene interaction) on different tomato cultivars (Alexander 1962). Verticillium dahliae race 1 contains an avirulence effector gene Ave1 that activates the tomato resistance gene (Ve1), which is present in most modern tomato cultivars (de Jonge et al. 2012). The Ave1 gene is absent in V. dahliae race 2 isolates, and these isolates can thus avoid recognition and are potentially virulent on all tomato cultivars. In South Africa, V. dahliae race 1 was first reported on tomatoes in 1977 (Gorter 1977), followed by the identification of race 2 in 1989 (Ferreira et al. 1990). Aside from utilising tomato plants containing and lacking the Ve1 gene for race identification, identification can also be conducted using race-specific PCR analyses (de Jonge et al. 2012; Short et al. 2014; Suaste-Dzul et al. 2023). Jiménez-Díaz et al. (2017) reported that the race-specific amplicon of V. dahliae race 2 isolates is polymorphic, and that isolates can based on the sequence genotypes, be divided into five haplotypes namely I, II, III, IV and V.
Verticillium dahliae is an anamorphic fungus and reproduces asexually, which mainly gives rise to a clonal population structure on the various hosts of the pathogen (Balloux et al. 2003). Genetic diversity can, however, be generated within asexual fungi via mutation, parasexuality, or somatic recombination (Giraud et al. 2008; Amyotte et al. 2012). Furthermore, genetic variation may also have been generated historically through sexual reproduction which was subsequently lost in modern-day agricultural populations (Milgroom et al. 2014). In V. dahliae populations, genetic diversity has traditionally been studied using vegetative compatibility grouping (VCG), which assesses the ability of the hyphae of paired isolates to fuse. Seven V. dahliae VCGs have been identified from various hosts and geographical localities including VCGs 1A, 1B, 2A, 2B, 4A, 4B and 6 (Jiménez- Díaz et al. 2006). Jiménez-Díaz et al. (2006) reported that diversity existed within VCG 2B based on conventional complementation studies and therefore two groups were designated within VCG 2B namely VCG 2Br and VCG 2Ba. The diversity within VCG 2B was supported by amplified fragment length polymorphism analysis (AFLPs) and sequence data from five genes, which resulted in the identification and designation of VCG 2B334 and 2B824 (Collado-Romero et al. 2006, 2008). Most recently, Milgroom et al. (2014), using a genome-based single-nucleotide polymorphisms (SNPs) analysis, further confirmed diversity within VCG 2B since three distinct lineages namely 2BR1, 2B334 and 2B824 were evident. Although VCG 1A and VCG 1B were first thought to belong to the same clonal lineage, it is now known that there is genetic differentiation among individuals belonging to VCG 1A and VCG 1B (Milgroom et al. 2016). Altogether, this has resulted in the designation of a total of nine V. dahliae clonal lineages (1A, B, 2A, 2BR1, 2B334, 2B824, 4A, 4B and 6) (Jiménez-Díaz et al. 2017). The lineages were named after their respective VCGs using conventional VCG testing, except for lineage 2BR1, which was not previously described (Milgroom et al. 2014).
The diversity within VCG 2B can be identified using a pathotype-genotype PCR approach. A pathotype-specific PCR assay for V. dahliae was first reported for cotton (Mercado-Blanco et al. 2001, 2002, 2003) where defoliating and nondefoliating pathotypes can be differentiated based on symptom development on inoculated plants (Schnathorst and Mathre 1966). Subsequently, Collins et al. (2005) reported the differential association of a 334 and 824 bp band from one of the pathotype-specific PCR assays with VCG 2B isolates. Collado-Romero et al. (2006) used this association along with their AFLP genotyping results to designate the VCG 2B isolates as lineages 2B334 or 2B824. Jiménez-Díaz et al. (2006) used the DNA banding patterns obtained from two multiplex pathotype-specific PCR assays to assign pathotype-genotypes (A, B, C and E) to V. dahliae isolates. Later, genotypes B and C were shown to belong to clonal lineages 2B334 and 2B824 respectively (Jiménez-Díaz et al. 2017).
Several studies have investigated the VCGs and races associated with V. dahliae isolates from tomato, but only a few have investigated the clonal lineages, pathotype-genotypes and race-haplotypes. Collado-Romero et al. (2008) investigated the pathotype-genotypes of two tomato isolates using one of the pathotype-genotype PCR assays. Milgroom et al. (2014) included two tomato isolates in their clonal lineage study. The study of Jiménez-Díaz et al. (2017), mainly included isolates from the study of Collado-Romero et al. (2006; 2008) and Milgroom et al. (2014), but also an additional five tomato isolates. Although the V. dahliae isolates in these studies were from multiple locations and hosts, no V. dahliae isolates were included from South Africa. In South Africa, apart from VCG testing conducted by Uys in 1996 (Uys 1996), no studies have been conducted on the genetic diversity of tomato V. dahliae isolates or any other host in South Africa.
Various inoculation methods have been used to assess the pathogenicity (ability to cause disease) and aggressiveness (degree of pathogenicity) of V. dahliae isolates on tomatoes and other hosts. The most frequently used methods involve the node-inoculation and root-dip methods (Ashworth 1983; Correll et al. 1988; Koike et al. 1994). No comprehensive study has been conducted on tomatoes to determine which inoculation method is best for determining the pathogenicity and aggressiveness of isolates. Reliable inoculation methods are important for successful aggressiveness and pathogenicity screening of Verticillium isolates on tomatoes. A reliable method should employ an adequate but not excess amount of inoculum that is evenly distributed upon inoculation to ensure that consistent results are obtained for all isolates in different experiments. Inoculation methods that are not labour intensive and that can be conducted in a short time frame can facilitate the screening of many Verticillium isolates and tomato accessions.
Studying the association between the genetic diversity within V. dahliae populations, pathogenicity and aggressiveness is key for the development of management strategies and screening tomato cultivars for resistance or tolerance to the pathogen. Therefore, the aim of this study was to characterise Verticillium isolates associated with Verticillium wilt of tomatoes in the Limpopo Province in South Africa. The characterisation involved the identification of the pathotype-genotypes, race-haplotypes and VCGs. Several inoculation methods were compared to identify a fast and reliable method for screening the pathogenicity of three V. dahliae isolates on tomato seedlings with and without the Ve1, and to investigate if isolates differed in aggressiveness. Subsequently, one of the inoculation methods was utilised to assess, under glasshouse conditions, the pathogenicity and aggressiveness of the genetically characterised Verticillium isolates.
Materials and methods
Verticillium wilt survey and isolation
Tomato plants showing symptoms of Verticillium wilt were collected in the Limpopo Province in the three major production regions (Highveld, Lowveld and Soutpansberg). Symptomatic plants were collected in 2013 from ten tomato fields, which included two fields in each of the Soutpansberg and Highveld regions, and six fields in the Lowveld. In each field, ten symptomatic plants were randomly selected within a one-hectare block. Sampling was conducted in two separate surveys during winter (Soutpansberg and Lowveld) and summer (Highveld) when the plants had reached maturity and were producing fruit (approximately 12 to 20 weeks post-plant).
Stems from the plants were washed in tap water and the crown area of each stem was cut into two 1 cm pieces. One piece was surface sterilized with 70% alcohol for 1 min and the other was rinsed in sterile distilled water. Each 1 cm piece was cut longitudinally with sterile secateurs, and small pieces (0.5 × 0.5 mm) of the vascular tissue were removed and placed on Petri dishes containing potato dextrose agar (PDA) (Biolab, Modderfontein, South Africa) and 0.05 g streptomycin (PDA+) or a modified NP-10 medium (Kabir et al. 2004). Dishes were incubated at room temperature for 2 weeks and resulting colonies were transferred to divided Petri dishes with PDA+ and carnation leaf agar (Fisher et al. 1982). Colonies with the characteristic morphology of Verticillium spp. (Inderbitzin et al. 2011) were identified using a light microscope and pure cultures were established from single spores. All isolates were maintained on potato carrot dextrose agar slants [extracts from 20 g potato and 20 g carrot pieces, 15 g biological agar (Biolab) and 0.05 g streptomycin in 1 L distilled water] at 4 °C. The forty V. dahliae isolates that were fully genetically characterised were deposited in the National Collection of Fungi (PPRI collection) housed at ARC-PHP in Pretoria, South Africa (Table 1).
Verticillium dahliae reference isolates
A set of nit mutant tester V. dahliae isolates previously characterised by Joaquim and Rowe (1990) namely V44 (clonal lineage 1 A, VCG 1A, PCR pathotype-genotype A, race 2 haplotype V), PH (VCG 2A), 115 (VCG 2B), BB (VCG 4A) and S39 (clonal lineage 4B, VCG 4B, PCR pathotype-genotype C, race 2 haplotype I) were used for VCG complementation studies and in selected PCR assays. The reference isolates were provided by Maria del Mar Jimenéz-Gasco (Department of Plant Pathology and Environmental Microbiology, The Pennsylvania State University, University Park, USA). Two of the isolates, V44 and S39, were used in the study of Jiménez-Díaz et al. (2017).
Species identification of Verticillium isolates
A collection of 262 Verticillium isolates obtained in the isolation studies were molecularly identified to the species level. Cultures were grown for 2 weeks at 22 °C on Petri dishes with PDA+ and carnation leaf agar. DNA was extracted using the Promega Wizard® SV Genomic DNA Purification System according to the manufacturer’s instructions. The exception was that at the start of the DNA extraction protocol, an additional step was included where 400 µl lysis buffer was added along with 2 mm glass beads into 2 ml tubes, which were shaken for 5 min at 30 l/s frequency using a Mixer Mill Type MM 301 (Retsch Gmbh, Haan, Germany).
The ITS region was targeted for identifying isolates as V. dahliae using V. dahliae species-specific primers Df and Dr (Inderbitzin et al. 2013), which yields a 490 bp amplicon. The PCR reaction (25 µl) consisted of 0.2 µM of each primer (Df and Dr), 1x Taq Master mix RED containing 1.5 mM MgCl2 (AMPLIQON, Odense, Denmark) and 2 µl template DNA. The PCR cycling conditions were: 2 min at 94 °C, followed by 32 cycles of 10 s at 94 °C, 20 s at 67 °C, 1 min at 72 °C and a final extension cycle of 7 min at 72 °C. Positive controls (isolates V44, PH and S39) and a negative control (no template DNA) were included to ensure that no contamination was present. All PCR reactions in this study were performed with a GeneAmp PCR System 9700 (Applied Biosystems, California, USA). Amplification products were visualised by separation on 1.5% agarose gels, stained with 0.5 µg/ml ethidium bromide and run in a 1 × TAE buffer (0.04 M Tris, 0.02 M glacial acetic acid and 1.27 mM EDTA, pH 7.85) followed by UV light illumination for visualisation. A 50 bp DNA size marker (HyperLadder™ 50 bp; Bioline, London, United Kingdom) was included in all agarose gels.
A subsample of four Verticillium isolates were selected from the isolates that produced the expected 490 bp V. dahliae amplicon, for which the ITS region was sequenced to further confirm the species identity. The post PCR clean-up was performed using a Nucleofast® 96 well PCR plate (Macherey-Nagel GmbH and Co., Düren, Germany) according to the manufacturer’s instructions. The sequencing reaction and cycle conditions were carried out as recommended by the manufacturer with a Big Dye® Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems). The resulting fragments were analysed on an ABI 3130xl Prism Genetic Analyzer (Applied Biosystems) by the Central Analytical Sequencing Facility (CAF) at Stellenbosch University. Sequences were aligned using the computer programme Geneious R10 (Biomatters Ltd., Auckland, New Zealand) and were BLAST analysed to reference strains in Genbank submitted by Inderbitzin et al. (2011).
VCG determination
For VCG testing, reference nit mutant isolates of VCG 1A, VCG 2A, VCG 2B, VCG 4A and VCG 4B were included (see reference isolate section). It was not possible to obtain reference nit mutant isolates for VCG 1B and VCG 6. This, however, was unlikely to have affected the analysis negatively since VCG 1B has only been isolated from woody plants and VCG 6 from peppers (Jiménez-Díaz et al. 2017). Forty V. dahliae tomato isolates (one isolate from each plant from which Verticillium was isolated) were characterised to the VCG level by first creating nit mutants, followed by the pairing of complementary nit mutants with the reference nit mutant isolates to determine heterokaryon formation.
Verticillium dahliae nit mutants were generated on water agar chlorate medium (WAC; 20 g of agar and 20 g sucrose amended with 20 g potassium chlorate per liter of distilled water) (Korolev and Katan 1997). The phenotype of nit mutants was determined on minimal agar medium (MM; Correll et al. 1987) amended with different nitrogen sources namely ammonium tartrate (Sigma-Aldrich, Kempton Park, South Africa) hypoxanthine (Sigma-Aldrich, Kempton Park, South Africa) and sodium nitrite (Labchem, Edenvale, South Africa). Nit mutants that were able to use all nitrogen sources except nitrate were phenotyped to nit 1, and nit mutants able to use all nitrogen sources except nitrate and hypoxanthine were phenotyped as nit M (Correll et al. 1987). Over 600 nit mutants were phenotyped for the 40 V. dahliae isolates (one isolate from each plant from which Veriticillium was isolated), from which four of each nit 1 and nit M mutants were selected per isolate. The four nit mutants (nit 1 and nit M) from each isolate were paired in every possible combination with the testers. Pairing was done by placing mycelial plugs of the nit mutants 1 cm apart on MM medium in 90 mm Petri dishes with the nit mutant of a known VCG in the middle. Cultures were incubated at 22 °C for 4 weeks and evaluated for heterokaryon formation.
Molecular genetic characterisation of isolates
PCR-based pathotype-genotype identification
The pathotype-genotype (defoliating or nondefoliating) was determined for the forty V. dahliae isolates, which were subjected to VCG testing, using two different sets of pathotype-specific PCR assays. The PCR band sizes obtained from the two sets of PCR assays were utilised to classify isolates as defoliating (D) or nondefoliating (ND) into four pathotype-genotypes (A, B, C and E) (Jiménez-Díaz et al. 2017). For the first pathotype-specific PCR assay primers DB19/DB22/espdef01 (Mercado-Blanco et al. 2003) were used in a multiplex PCR reaction (25 µl), which contained each primer (DB19/DB22/espdef01) at a concentration of 0.7, 0.1 and 0.2 µM respectively along with 1x Taq Master mix RED (1.5 mM MgCl2) and 2 µl template DNA. The PCR cycling conditions were: 4 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min at 60 °C, 30 s at 72 °C and a final extension cycle of 6 min at 72 °C. The DB19/DB22/espdef01 primers yield a 523 bp amplicon for the nondefoliating (ND) pathotype and 539 bp and 334 bp amplicons for the defoliating (D) pathotype (Jiménez-Díaz et al. 2006). The second pathotype-specific assay consisted of a multiplex assay with primers INTD2f/INTD2R/INTND2f/INTND2r (Mercado-Blanco et al. 2001, 2002). Within the multiplex reaction, the primers INTD2f/INTD2R and INTND2f/INTND2r produce an amplicon of 462 or 824 bp, respectively (Jiménez-Díaz et al. 2006). The same PCR reaction mixture and cycling conditions were used as described for the first pathotype assay, except that each primer pair was added at a concentration of 0.2 µM, the annealing temperature was set at 64 °C and the elongation step was increased to 1 min. Reactions for both pathotype assays included a negative control (no template DNA) and positive controls for the D (isolate V44) and ND (isolate S39) pathotypes. The presence and absence of the specific amplicons in the two pathotype-specific assays were used to determine the pathotype-genotype (A, B, C or E) of the V. dahliae isolates: pathotype-genotype A = 334 bp (+), 462 bp (+), 824 bp (-); B = 334 bp (+), 462 bp (-), 824 bp (-); C = 334 bp (-), 462 bp (-), 824 bp (+); E = 334 bp (+), 462 bp (-), 824 bp (+) (Jiménez-Díaz et al. 2017).
PCR-based race determination
The 40 isolates that were characterised for pathotype-genotype, were also assessed for their race identity (race 1 or race 2) using two PCR assays that are each specific to either one of the two races. Primer pair VdAve1F/VdAve1R (de Jonge et al. 2012) amplifies race 1 and produces a 900 bp amplicon for race 1 isolates, and no amplicon for race 2 isolates. Primer pair VdR2F/VdR2R (Short et al. 2014) amplifies race 2, where it produces a 256 bp amplicon; no amplicon is produced for race 1 isolates. PCR reactions consisted of the same reaction mixture as for the pathotype-genotype PCR assays except for the primer concentrations, which consisted of 0.4 µM of each primer for both assays. The PCR cycling conditions were: 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 62 °C (race 1 primers) or 64 °C (race 2 primers), 30 s at 72 °C and a final extension cycle of 7 min at 72 °C. Reactions included a negative control (no template DNA) and positive controls for race 1 (isolate PH) and race 2 (isolate S39).
The presence of different race haplotypes (I to V) within race 2 was determined through sequencing of the 256 bp amplicons obtained in the race 2-specific PCR assay (Jiménez-Díaz et al. 2017). The amplicons were excised from the agarose gel and purified using the Promega Wizard® SV Gel and PCR Clean-Up system. Amplicon sequencing was conducted by CAF as described in the species identification section.
Clonal lineage identification
The results from the VCG testing and the pathotype-genotype analyses of the isolates were used to determine the clonal lineage using the approach of Jiménez-Díaz et al. (2017).
Comparison of inoculation methods for assessing pathogenicity and aggressiveness
Seven different inoculation methods were tested namely the agar plug (Koike et al. 1994), millet (Strauss and Labuschagne 1995), node-inoculation (Ashworth 1983), root-dip (Koike et al. 1994), sand-bran (Lamprecht 1986), soil-drench (Qin et al. 2008) and toothpick (Hormchong 1969) methods.
Plants
Tomato seeds of the cultivar Heinz 1370 (without Ve gene, susceptible to races 1 and 2) were sown individually into 2 × 2 × 10 cm (length x width x depth) square punnets filled with a pasteurised peat-based growth medium (Hygromix, Hygrotech, Stellenbosch), which were held in seedling trays. Seedlings were maintained on benches in a glasshouse at ± 25 °C day and ± 18 °C night temperatures for 4 weeks. After 4 weeks, seedlings were subjected to different inoculation treatments. For the node-inoculation, soil-drench and toothpick methods the seedlings were transplanted into 18 cm pots, three seedlings per pot, the day before treatment. The seedlings utilised for the sand-bran and millet seed were transplanted the day after treatment and for the root-dip method, the seedlings were transplanted on the day of treatment, also three seedlings per 18 cm pot. For all treatments, seedlings were transplanted into pasteurised (30 min at 82 °C) Hygromix. Pots were incubated in a glasshouse at ± 18 °C night and ± 25 °C day temperatures.
Isolates
Three V. dahliae single-spore isolates (T73, T83, T85) (Table 1) were grown for 2 weeks at 22 °C on divided Petri dishes with PDA+ and carnation leaf agar (a preliminary study indicated these isolates to be pathogenic on tomatoes; data not shown). All three isolates were VCG 4B, pathotype-genotype C and race-haplotype I. For each inoculation method, three pots were inoculated (three plants per pot) for each of the three V. dahliae isolates. Pots were arranged in a randomised block design on glasshouse benches and the trial was repeated once.
Inoculation methods
Agar plug method. The agar plug method was utilised as described by Koike et al. (1994), with a slight modification. A 5 mm2 agar plug was used instead of a 3 cm2 size agar plug. The agar plug (5 mm2) of each isolate was transferred from the divided dish (PDA+ and carnation leaf agar) to the centre of a 90 mm Petri dish containing PDA+. The plates were incubated for 2 weeks at 22 °C. The inoculation of tomato seedlings consisted of placing a colonised 5 mm PDA+ agar plug into each planting hole before transplanting each tomato seedling. Control treatments consisted of a 5 mm PDA+ agar plug without any fungal growth.
Millet seed method. The millet inoculum was prepared according to the method described by Strauss and Labuschagne (1995). The inoculum consisted of 200 g millet seeds that were soaked in 100 ml water, in a 1 L Schott bottle for 12 h after which the excess water was drained. Each bottle was autoclaved for two consecutive days at 120 °C for 20 min and inoculated on the third day with 10 PDA+ plugs (± 1 cm). The control treatment was inoculated with uncolonised agar plugs. The inoculum was incubated for 11 days at 22 °C in the dark and shaken every third day to ensure proper colonisation. The colonised millet was mixed with the Hygromix at a concentration of 0.05% (w/w) in 18 cm diameter pots, 24 h before planting.
Node-inoculation method. The node-inoculation method used by Ashworth (1983) was utilised. Fresh cultures were prepared on divided Petri dishes containing PDA+ and carnation leaf agar and grown for 2 weeks. Conidia were harvested from the plates using distilled water and a conidial suspension of 1 × 106 conidia/ml was prepared for each V. dahliae isolate on the day of inoculation. Spore suspensions were counted using a haemacytometer and a light microscope. Four-week-old plants were inoculated by injecting the spore suspension once, using a 1.80-mm-diameter (15 gauge) needle, at the internode immediately below the apical meristem. Approximately 1 ml of the spore suspension was inoculated per plant. The control treatment consisted of sterile distilled water.
Root-dip method. The root-dip inoculation method was utilised as described by Correll et al. (1988), with a slight modification since the roots were not clipped before being dipped into the spore suspension. A spore suspension was prepared as described above for the node-inoculation method. Four-week-old seedlings were dipped in the conidial suspension for five minutes and were transplanted into pots following the inoculation. The control treatment consisted of sterile distilled water.
Sand-bran method. The sand-bran inoculum was prepared according to the method described by Lamprecht (1986). The inoculum consisted of 400 g river sand and 20 g wheat bran that were mixed in a 1 L Schott bottle with 60 ml sterile distilled water. Each bottle was autoclaved for three consecutive days at 120 °C for 1 h on the first day and 30 min on the last two days. On the fourth day, the sand-bran was inoculated with 10 PDA+ plugs (5 mm in diameter) of each V. dahliae isolate. The control treatment was inoculated with clean agar plugs. The inoculum was incubated for 11days at 22 °C in the dark and shaken every third day to ensure proper colonisation. The sand-bran inoculum was applied at a concentration of 0.05% (w/w) to the pots as described for the millet seed inoculation method.
Soil-drench method. The soil-drench inoculation method was utilised as described by Qin et al. (2008), with modifications. The seedlings were not drenched with the spore suspension before transplanting but post-transplant, and the volume of conidial suspension was increased from 1.5 ml to 75 ml. A spore suspension was prepared for the soil-drench inoculation method as described for the node-inoculation method. Pots, containing, 4-week-old plants were drenched with 75 ml of the conidial suspension, 24hours after transplanting the seedlings. The control treatment consisted of sterile distilled water.
Toothpick method. The toothpick inoculation method of Hormchong (1969) was used, but it was modified so that the stem was inoculated with the toothpicks and not the petiole. Toothpicks were sterilised in distilled water on two consecutive days. Sterile toothpicks were placed in a circle around a 5 mm agar plug of each V. dahliae isolate, on a Petri dish containing PDA+, which were colonised during the 2 weeks of incubation. The colonised toothpicks were used to pierce the stem of each tomato seedling (approximately 1 cm above soil level), 24 h post-transplant of the 4-week-old seedlings. The control treatment involved piercing the stem with sterile toothpicks.
Evaluation of the inoculation methods
Infection was confirmed through isolations from the stems of all the inoculated and control plants onto NP-10 medium as described above. Each dish was examined microscopically 5 days after plating for the presence of V. dahliae and dishes were screened up until 28 days post-plant.
Plant height and disease severity was assessed 4 weeks after inoculation. Increase in plant height was calculated using the plant height determined at the start and end of the trial. The height was measured from the first node of the main stem (where the first two seed leaves developed) to the dominant growth tip. Increase in plant height was averaged per pot. Disease severity was assessed using the foliar symptom range of 0–6 described by Iglesias-Garcia et al. (2013); 0 = no symptoms, 1 = > 0 to 10% of the plant showing chlorosis or necrosis, 2 = > 10 to 25%, 3 = > 25–50%, 4 = > 50–75%, 5 = > 75–100% and 6 = dead plant. Disease severity was converted to percentage values (midpoints). Midpoints of the severity ratings were then averaged per pot.
Statistical analysis of inoculation methods
Levene’s ratio test was performed to test for homogeneity of trial variances between the trial repeats (Levene 1960). Data from the two independent trials were considered block treatments providing that Levene’s variance ratio test showed homogeneity in trial variance. Disease severity and increase in plant height data were subjected to analysis of variance (ANOVA) using the GLM (General Linear Models) procedure of SAS statistical software (version 9.2; SAS Institute Inc.). Within the GLM the block and trial was set as random effects. A factorial ANOVA analysis (trial repeat, isolate and treatment) was conducted for both data sets. The Shapiro-Wilk test was performed to test for normality (Shapiro and Wilk 1965). In cases where deviations from normality were due to kurtosis and not skewness, the data were accepted as reliable and the results were interpreted without transformation (Glass et al. 1972). The Fisher’s t-test was calculated at the 95% confidence level to compare treatment means for significant effects.
Evaluation of the aggressiveness of isolates
Plants and isolate inoculation
This experiment was performed to determine whether the aggressiveness of the 40 V. dahliae differed on the cultivar Floradade that contains the Ve gene (i.e. susceptible only to race 2, but resistant to race 1). Cultivar Floradade was used since at the time of the experiment, all the isolates were known to belong to race 2 based on the PCR race-specific results (see race-specific PCR results). The sand-bran inoculation method was used as described above. For this purpose, 40 single-spore V. dahliae race 2 isolates were grown for 2 weeks at 22 °C on divided Petri dishes with PDA+ and carnation leaf agar. The sand-bran inoculum was prepared for each isolate and the control treatment as described above. Seedlings of the cultivar Floradade were prepared as previously described and were transplanted into pots containing the sand-bran inoculated Hygromix. Three plants were planted per 18 cm pot and three repeats were planted per isolate. Pots were incubated in a glasshouse at 18 °C night and 25 °C day temperatures. Pots were arranged in a completely randomised block design on glasshouse benches and the trial was repeated once. Increase in plant height and disease severity were evaluated and analysed as described for the inoculation method experiments.
Statistical analysis
The statistical analyses were conducted as described for the inoculation methods.
Results
Species identification of Verticillium isolates
A total of 262 Verticillium isolates were obtained from nine of the ten sampled fields. Verticillium dahliae was isolated from the Highveld (two fields, 30% of the plants), Lowveld (six fields, 67.5% of the plants) and Soutpansberg area (one field, 2.5% of the plants). The V. dahliae-specific PCR assay along with the two V. dahliae reference isolates all yielded the 490 bp V. dahliae-specific amplicon. The ITS sequences of the four analysed isolates (accessions OQ612473 -OQ612476) all had 99% identity to the V. dahliae GenBank accession/s of Inderbitzin et al. (2011).
VCG determination
Based on heterokaryon formation with the VCG reference strains 38 isolates were assigned to VCG 4B and two isolates (T61 and T62) to VCG 2B. Isolates T61 and T62 were from the Lowveld region (Table 1). The two VCG 2B isolates had a pathotype-genotype C (see below), and thus belonged to clonal lineage VCG 2B824.
Molecular genetic characterisation of isolates
PCR-based pathotype-genotype identification
The two pathotype-genotype specific multiplex PCR’s yielded a 523 and 824 bp amplicon, respectively, for all the V. dahliae isolates (Fig. 1). Therefore, the combination of the two sets of PCR assays identified all 40 V. dahliae isolates as pathotype-genotype C (Table 1).
Gel electrophoresis of PCR amplicons to identify defoliating (D) and nondefoliating pathotypes in Verticillum dahliae and for determining pathotype-genotypes (A, B, C and E; (Jiménez-Díaz et al. 2017). The presence or absence of specific amplicons in two PCR assays that can potentially generate (A) 334 and 523 bp amplicons with primers DB19/DB22/espdef01 and (B) 824 or 462 bp amplicons with primers INTD2f/INTD2R/INTND2f/INTND2r were used to identify the South African V. dahliae tomato isolates (T2, T21, T32, T55, T61, T62, T71, T88 and T91) as nondefoliating (+ 523 bp) pathotype-genotype C [334 bp (-), 462 bp (-), 824 bp (+)]. The international reference isolates for non-defoliating pathotype-genotypes B [334 bp (+), 462 bp (-), 824 bp (-)] and C were isolates V44 and S39 respectively. A 50-bp ladder was loaded in the first and last lane of the gels
PCR-based race determination
The race-specific PCR-based analysis only yielded a 256 bp amplicon for all isolates, which thus identified all 40 V. dahliae isolates as race 2 (Fig. 2). Based on sequence data of the race 2 amplicon of the two VCG 2B and 38 VCG 4B isolates, the two VCGs were identified as haplotype II and I, respectively (Table 1).
Gel electrophoresis of PCR amplifications to identify race 1 and race 2 in isolates of Verticillium dahliae. (A) Race 1-specific 900-bp amplicon generated with primers VdAve1F and VdAve1R. (B) Race 2-specific 256-bp amplicons generated with primers VdR2F and VdR2R. Reactions included a negative control (no template DNA) and positive control for race 1 (PH) and race 2 (S39). A 50 bp DNA ladder was loaded in the first and last lane of the gels
Clonal lineage identification
The clonal lineage of the isolates was determined based on the VCG since the clonal lineages of Milgroom et al. (2014) in almost all cases correspond to VCG (Jiménez-Díaz et al. 2012; Jiménez-Gasco et al. 2014; Milgroom et al. 2014). The exception is for VCG 2B since three lineages are present (2B334, 2B824 and 2BR1). However, two of the lineages can be identified based on pathotype-genotype PCR results since genotypes B and C belong to lineages 2B334 and 2B824 respectively (Jiménez-Díaz et al. 2017). Thirty-eight of the V. dahliae isolates thus belonged to clonal lineage 4B and two isolates (T61 and T62) to clonal lineage 2B824 (Table 1).
Comparison of different inoculation methods for assessing pathogenicity and aggressiveness
Four weeks post-inoculation, symptoms of chlorosis and necrosis became prominent (Fig. 3), although at different severity levels for the different treatments. Control plants for all seven inoculation methods remained symptomless. Verticillium dahliae could be re-isolated from the plants from all seven methods and no V. dahliae was isolated from the control plants.
Levene’s test indicated homogeneity of data for disease severity (P = 0.7290) for the two trial repeats. For disease severity, a significant trial x isolate x treatment interaction was observed (P = 0.0017) (Table 2). This interaction was difficult to investigate due to the complexity of the t-test results. Since there was no significant trial x treatment (P = 0.7244) or trial x isolate (P = 0.0621) interactions, the interaction could be attributed to the significant isolate x treatment (P = 0.0006) interaction (Table 2). The isolate x treatment interaction (Table 3) was thus investigated for disease severity since it showed that the isolates responded differently to the treatments (inoculation methods).
The disease severity results showed that for all three isolates the toothpick and sand-bran methods resulted in significantly higher disease severity levels than most of the other inoculation methods (Table 3). The exception was for the root-dip method where isolates T83 and T85, but not T73, yielded disease severity levels that did not differ significantly from the sand-bran and toothpick methods. Inoculation methods that did not differ significantly from the root-dip method in disease severity levels included the millet (isolate T73), node-inoculation (isolates T73 and T85) and soil drench (isolate T73) methods.
Using disease severity as a parameter, the aggressiveness of the three isolates sometimes differed significantly but this was inoculation method dependent. Based on the toothpick and node-inoculation methods the three isolates did not differ significantly in disease severity. The sand-bran and root-dip methods differed in identifying the relative aggressiveness of the isolates but were comparable in showing differences in aggressiveness. The sand-bran inoculation method showed that isolate T85 was more aggressive than isolate T83, with isolates T73 and T83 not differing in aggressiveness. The root-dip method resulted in isolate T83 being more aggressive than T73, whereas isolates T83 and T85 did not differ significantly in aggressiveness. In addition to the sand-brand, root-dip and toothpick methods, the node-inoculation method was the only method that identified all three isolates as pathogenic. The agar method was ineffective as an inoculation method since it indicated that all three isolates were non-pathogenic, whereas the millet and the soil-drench inoculations could not identify all three isolates as pathogenic.
For the increase in plant height, Levene’s test was non-significant (P = 0.2274) for the trial repeats. There was no significant trial x isolate x treatment (P = 0.4960), trial x treatment (P = 0.1301) or trial x isolate (P = 0.5293) interactions for plant height, and therefore the significant isolate x treatment (P < 0.0001) interaction (Table 2) was investigated (Table 3).
The inoculation methods resulted in significantly different plant heights for the uninoculated control plants (Table 3). The agar plug method resulted in a significantly higher plant height than the other methods, except for the toothpick method. Furthermore, the root-dip method control plants had significantly higher plant heights than the sand-bran method control plants. Because of the effect of the inoculation method on plant height, the effect of the inoculated isolates was investigated for each isolate relative to the relevant uninoculated control inoculation method.
Similar to disease severity, the inoculation methods differed in identifying isolates as pathogenic, and with regards to the relative aggressiveness of isolates (Table 3). The agar plug, soil-drench, sand-bran and millet seed methods did not result in significant differences in plant heights relative to the relevant control plants for all three isolates, i.e. the isolates were non-pathogenic based on this parameter. The node-inoculation method only identified isolate T85 as pathogenic (significantly lower plant height than the uninoculated control node-inoculation). The toothpick inoculation method yielded a significant reduction in plant heights for isolates T85 and T73 relative to the toothpick control plants, with isolates not differing significantly from each other in aggressiveness. The root-dip method was the only method that enabled all three isolates to cause a significant reduction in plant heights relative to the control root-dip plants. The three isolates did not differ significantly in aggressiveness (plant height) when inoculated as a root-dip.
Evaluation of the aggressiveness of isolates
There was homogeneity in the disease severity (P = 0.3897) and increase in plant height (P = 0.7283) data for the two trial repeats based on Levene’s test for the glasshouse trial. ANOVA showed that there was no significant trial x treatment interaction for disease severity (P = 0.4149) and increase in plant height (P = 0.2260), and the repeated trial data were thus combined for each of the parameters. There was a significant difference among isolates in disease severity (P < 0.0001) and plant height (P = 0.0121).
Thirty-two of the 40 V. dahliae isolates were considered pathogenic towards Floradade since the isolates caused disease severities that differed significantly from the control (Table 4). Isolates T2, T3, T73, T74, T75, T83, T84 and T91 were non-pathogenic toward Floradade since the incited disease severities did not differ significantly from the control. Two of these isolates (T73 and T83) were, however pathogenic on Heinz in the experiment where different inoculation methods were evaluated. Isolates T22, T23 and T94 were more aggressive than 11 of the other isolates (T31, T34, T43, T52, T61, T62, T64, T82, T93, T95 and T96) since the isolates caused significantly higher disease severity ratings.
For increase in plant height, none of the isolates were able to significantly reduce plant height when compared with the control plants (Table 4). Inoculation with isolate T62 and T85 resulted in a significantly higher plant height than the control. Verticillium dahliae was recovered from plants inoculated with each respective isolate, which confirmed infection. No V. dahliae was isolated from the control plants.
Discussion
In the current study, Verticillium dahliae was identified as the only Verticillium species associated with Verticillium wilt of tomato in the Limpopo Province in South Africa. The population was clonal based on the investigated markers since only two VCGs (2B and 4B), two race-haplotypes (I and II) and one pathotype-genotype (C) were identified among 40 isolates. The VCG and pathotype-genotype data allowed to establish that the isolates belonged to clonal lineages 4B and 2B824. The most aggressive isolates were identified within lineage 4B, but since only two isolates belonged to lineage 2B824 limited conclusions could be made regarding the relative aggressiveness of the two lineages.
Internationally, the VCGs associated with tomatoes have been identified as VCG 2A, VCG 2B, VCG 4B. Genetic characterisation of the V. dahliae isolates from tomatoes sampled at nine of the locations surveyed in South Africa in the Limpopo Province showed that VCG 4B (corresponding to lineage 4B) was widespread (95% of isolates) occurring in all three tomato production regions whereas VCG 2B (corresponding to lineage 2B824) had a limited distribution (5% of isolates) and was only identified in the Lowveld. Other studies have also reported that VCG 4B is the dominant VCG in tomatoes. In Greece, among the 15 investigated isolates VCG 4B (73.34%) dominated with VCG 2B (13.33%) being less prevalent along with VCG 2A (13.33%) (Papaioannou et al. 2013). In Israel, VCG 2A (40.62%) and VCG 4B (53.13%) dominated, with VCG 2B (6.25%) also having a limited distribution among the 32 investigated isolates (Korolev et al. 2000). In Canada, three characterised tomato isolates were identified as VCG 2A, VCG 2B and VCG 4B (Dobinson et al. 1998). The studies in Greece, Israel and Canada did not identify the lineages present within VCG 2B. Among 10 tomato isolates from Israel, Spain and the USA that were investigated by Jiménez-Díaz et al. (2017) VCG 2A (60%, lineage 2A), VCG 2B (30%, lineage 2B824) and VCG 4B (10%, lineage 4B) were also identified as the only VCGs associated with tomato.
The race-specific PCR assay showed that all the South African V. dahliae isolates belonged to race 2. This was expected as most tomato cultivars, including cultivars Topasia and ZZX65 from which the isolations were conducted, contain the Ve resistance gene for V. dahliae race 1. In other countries, race 2 has also mainly been identified on tomatoes (Grogan et al. 1979; Jabnoun-Khiareddine et al. 2007; Short et al. 2014, Suast-Dzul et al. 2023).
The South African tomato V. dahliae isolates were all from the nondefoliating pathotype-genotype C and race-haplotype I or II, which is similar to the findings reported by Jiménez-Díaz et al. (2017). The nondefoliating pathotype phenotype corresponds with the symptoms observed in the field and glasshouse as no severe defoliation was observed. The South African race 2 tomato isolates belonged to haplotypes I (lineage 4B) and II (lineage 2B824) similar to what was reported by Jiménez-Díaz et al. (2017).
The root-dip method has been used in most studies involving pathogenicity testing of V. dahliae on tomatoes (Bender and Shoemaker 1984; Correll et al. 1988; O’Garro and Clarkson 1988; Ferreira et al. 1990; Baergen et al. 1993; Subbarao et al. 1995; Nagao et al. 1997; Uys et al. 1997; Bhat and Subbarao 1999; Kim et al. 2001; Tsror et al. 2001; Jabnoun-Khiareddine et al. 2007; Ligoxigakis and Markakis 2012; Iglesias-Garcia et al. 2013; Papaioannou et al. 2013; Jiménez-Díaz et al. 2017; Usami et al. 2017; Suaste-Dzul et al. 2022). On horseradish, the root-dip method was also identified as providing more consistent results than inoculations conducted with colonised oat seed or silica sand inoculum (Atibalentja and Eastburn 1997. Similarly, on cauliflower, the root-dip method was identified as the best of the four investigated inoculation methods (Koike et al.1994). Our study supports the use of the root-dip method for assessing pathogenicity and aggressiveness of V. dahliae on tomatoes, since the root-dip method was the only method out of the seven evaluated that resulted in a significant reduction in plant height for all three of the investigated isolates on cultivar Heinz. Disease severity ratings resulting from the root-dip method supported the plant height data. Disease severity rating can be subjective, and therefore plant height is useful as an unbiased parameter to assess the pathogenicity and aggressiveness of isolates. Other unbiased disease severity parameters that have been assessed for V. dahliae infected tomato plants, which may also be useful for assessing aggressiveness, include vascular discoloration (Tabaeizadeh et al. 1999; Bhat et al. 2003) and the number of symptomatic leaves per plant (Bhat et al. 2003). Vascular discoloration can be assessed by measuring the length of vascular discoloration at the base of plants, which can be converted to an index ([length of discoloration ÷ total plant length] × 100]) (Tabaeizadeh et al. 1999), determining the percentage vascular discoloration on a scale of 0 to 5, or the proportion of plants with vascular discoloration (Bhat et al. 2003). The root-dip method revealed significant differences in the aggressiveness of the isolates based on foliar disease severity but not for plant height. A draw-back of the root-dip method is that when many isolates must be tested, as was the case in some of our experiments, the labour and time involved in counting spore suspensions makes it challenging to use root-dip inoculations.
This study is the first study on tomatoes comparing several inoculation methods with each other. The second-best inoculation method for assessing pathogenicity and aggressiveness in this study was the sand-bran method. Although the sand-bran method did not affect plant height for the three inoculated isolates, it identified all three isolates as pathogenic based on disease severity ratings. Some of the isolates furthermore differed significantly in aggressiveness (significantly different disease severities) when inoculated using sand-bran. Although the toothpick method was also among the methods that yielded the highest disease severity ratings and it resulted in a significant reduction in plant height for two isolates, the method is not recommended since it was unable to reveal significant differences in disease severity and thus the aggressiveness of the three isolates. This is likely because the method does not assess the ability of isolates to overcome root defense responses (preformed and induced) and colonisation of roots prior to entering the stem vascular systems. These aspects likely form an important component of the aggressiveness of isolates. A similar problem will be associated with the node-inoculation method, which was the only other method that was able to identify all three isolates as pathogenic.
In this current study, the soil-drench, agar plug and millet methods were inconsistent in identifying isolates as pathogenic and these methods are thus a poor choice for pathogenicity testing on tomatoes. The poor performance of the agar plug method and soil-drench was also reported by other studies. Koike et al. (1994) compared four inoculation methods for pathogenicity testing of V. dahliae on cauliflower. The agar plug method was unable to cause a significant difference in disease severity on cauliflower when compared to the control plants (Koike et al. 1994). On cocoa plants where the soil-drench method was compared with a stem puncture method, the incubation time required for symptom development post-inoculation was three times longer than for the soil-drench method. The stem-puncture method furthermore caused significantly higher disease severities when compared to the soil-drench method (Leon-Ttacca et al. 2019). Since incubation time may influence the outcome of pathogenicity testing, it is possible that in our current study the soil-drench method if conducted for a longer period may have identified all three isolates as pathogenic (significant disease severities).
The trial evaluating the aggressiveness of 40 V. dahliae isolates towards the Floradade cultivar using the sand-bran inoculation method showed that only disease severity ratings, but not plant height assessments were able to identify some isolates as pathogenic. Our inoculations on Heinz 1370 (three isolates) also showed that sand-bran inoculations do not result in a significant reduction in plant height, only disease severity is significantly affected. Other tomato studies have also reported that plant height is a less accurate measurement of pathogenicity and aggressiveness (Visser and Hattingh 1981; Bender and Shoemaker 1984; O’Garro and Clarkson 1988; Tsror et al. 2001; Bhat et al. 2003), but that vascular discoloration may be useful (Tabaeizadeh et al. 1999; Bhat et al. 2003). In our study, the root-dip inoculation method, but not the sand-bran method, consistently reduced plant height on Heinz 1370. This is likely due to infections occurring faster with the root-dip method due to the large number of conidia loaded directly onto roots thus causing rapid infection and colonisation of the roots and consequently plant height inhibition. A similar phenomenon likely also explains why the node-inoculation and toothpick methods, although inconsistent for the three isolates, were the only other methods that significantly reduced plant height. Therefore, to obtain plant height reductions for V. dahliae on tomatoes in pot trials, high inoculum exposure or invasive inoculation techniques are required. On other hosts, parameters that have been used to reliably assess the aggressiveness of isolates also include disease severity ratings, in addition to a reduction in plant biomass and plant height (Xiao and Subbarao 1998; Bhat and Subbarao 1999; Villarroel-Zeballos et al. 2012).
The sand-bran method was inaccurate for evaluating the pathogenicity of the 40 genetically characterised V. dahliae isolates on cultivar Floradade (resistance to race 1 isolates), but was useful for evaluating the aggressiveness of the isolates. The method was inaccurate for pathogenicity identification since seven of the isolates were non-pathogenic on Floradade based on foliar disease severity ratings, whereas the race-specific PCR showed that the isolates were race 2. Assessment of vascular discoloration may have revealed the isolates as pathogenic and should be investigated in future. The inefficacy of the sand-bran method for evaluating pathogenicity on Floradade was unexpected since the method was suitable for evaluating the pathogenicity of all three investigated isolates on Heinz 1370. These included two of the isolates (T73 and T83) that were non-pathogenic on Floradade. This could reflect differences in minor genes conveying tolerance in the two cultivars, with Floradade being more tolerant than Heinz 1370. We selected the sand-bran method for evaluating the aggressiveness of isolates due to the time-consuming nature of the root-dip inoculation method that requires the counting of spore suspensions for 40 isolates that had to be inoculated on the same day. Although inaccurate for evaluating pathogenicity, the sand-bran method was able to identify three isolates as being more aggressive than 11 of the isolates with the lowest disease severity ratings. The three most aggressive isolates belonged to clonal lineage 4B. The higher aggressiveness of the three isolates should be confirmed in future studies using die root-dip inoculation method, since our study did not access whether the isolates colonised the sand-bran to a similar manner. Some isolates may be more aggressive colonisers of the sand-bran, which could affect their relative aggressiveness. Variability in aggressiveness among tomato V. dahliae isolates has also been reported by Jabnoun-Khiareddine et al. (2007).
In summary, V. dahliae is the only species associated with tomatoes in the Limpopo Province in South Africa where the pathogen has a clonal structure, with clonal lineage 4B dominating and 2B824 having a limited distribution. Our study has contributed significantly to our knowledge of the genetic characteristics associated with tomato V. dahliae isolates since only a few studies have been conducted worldwide. Future studies in South Africa should assess more isolates from the Highveld and Soutpansberg regions to ascertain that clonal lineage 2B824 does not occur in these regions. Fewer fields were surveyed in these two regions than in the Lowveld, which may have contributed to this clonal lineage only being identified in the Lowveld. No studies have been conducted to determine whether the higher aggressiveness of lineage 4B isolates contributes to its wider distribution. It will thus be important to investigate this in future studies by including more lineage 2B824 isolates in aggressiveness studies since only two were included in the current study, whereas 38 lineage 4B isolates were evaluated. The root-dip method is recommended for assessing the pathogenicity and aggressiveness of V. dahliae isolates, although it is logistically difficult to utilise when evaluating large numbers of isolates. The toothpick method may have the potential as a quick method for assessing pathogenicity, but it is unable to reveal differences in aggressiveness among isolates. The reliability of the toothpick method will have to be assessed further using a larger number of isolates to determine whether all the isolates will be identified as pathogenic on Floradade. It may be that similar to the sand-bran method, the toothpick method may be less effective on Floradade than Heinz. We have shown that the sand-bran method can be used for identifying three isolates that were more aggressive than 11 other isolates. This information along with the genotyping data should be taken into consideration for selecting isolates for future studies investigating effective management strategies and screening tomato cultivars for tolerance against V. dahliae.
Data Availability
Datasets are available from the corresponding author upon reasonable request.
References
Alexander LJ (1962) Susceptibility of certain verticillium-resistant tomato varieties to an Ohio isolate of pathogen. Phytopathology 52(10):998–1000
Amyotte SG, Tan X, Pennerman K, Jiménez-Gasco MM, Klosterman SJ, Ma L et al (2012) Transposable elements in phytopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification. BMC Genomics 13:314. https://doi.org/10.1186/1471-2164-13-314
Ashworth LJ (1983) Aggressiveness of random isolates of Verticillium dahliae from cotton and the quantitative relationship of internal inoculum to defoliation. Phytopathology 73:1292–1295. https://doi.org/10.1094/Phyto-73-1292
Atibalentja N, Eastburn DM (1997) Evaluation of inoculation methods for screening horseradish cultivars for resistance to Verticillium dahliae. Plant Dis 81(4):356–362. https://doi.org/10.1094/PDIS.1997.81.4.356
Baergen KD, Heweitt JD, St. Clair DA (1993) Resistance of tomato genotypes to four isolates of Verticillium dahliae race 2. HortScience 28(8):833–836. https://doi.org/10.21273/HORTSCI
Balloux F, Lehmann L, De Meeûs T (2003) The population genetics of clonal and partially clonal diploids. Genetics 164(4):1635–1644. https://doi.org/10.1093/genetics/164.4.1635
Bender CG, Shoemaker PB (1984) Prevalence of Verticillium wilt of tomato and virulence of Verticillium dahliae race 1 and race 2 isolates in western North Carolina. Plant Dis 68(4):305–309. https://doi.org/10.1094/PD-69-305
Bhat RG, Subbarao KV (1999) Host range specificity in Verticillium dahliae. Phytopathology 89(12):1218–1225. https://doi.org/10.1094/PHYTO.1999.89.12.1218
Bhat RG, Smith RF, Koike ST, Wu BM, Subbarao KV (2003) Characterization of Verticillium dahliae isolates and wilt epidemics of pepper. Plant Dis 87:789–797. https://doi.org/10.1094/PDIS.2003.87.7.789
Collado-Romero M, Mercado-Blanco J, Olivares-García C, Valverde-Corredor A, Jiménez-Díaz RM (2006) Molecular variability within and among Verticillium dahliae vegetative compatibility groups determined by fluorescent amplified fragment length polymorphism and polymerase chain reaction markers. Phytopathology 96(5):485–495. https://doi.org/10.1094/PHYTO-96-0485
Collado-Romero M, Mercado-Blanco J, Olivares-García C, Jiménez-Díaz RM (2008) Phylogenetic analysis of Verticillium dahliae vegetative compatibility groups. Phytopathology 98(9):1019–1028. https://doi.org/10.1094/PHYTO-98-9-1019
Collins A, Mercado-Blanco J, Jiménez-Díaz RM, Olivares C, Clewes E, Barbara DJ (2005) Correlation of molecular markers and biological properties in Verticillium dahliae and the possible origins of some isolates. Plant Pathol 54(4):549–557. https://doi.org/10.1111/j.1365-3059.2005.01240.x
Correll JC, Klittich CJR, Leslie JF (1987) Nitrate non-utilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77(12):1640–1646. https://doi.org/10.1094/Phyto-77-1640
Correll JC, Gordon TR, McCain AH (1988) Vegetative compatibility and pathogenicity of Verticillium albo-atrum. Phytopathology 78(8):1017–1021. https://doi.org/10.1094/Phyto-78-1017
de Jonge R, van Esse HP, Maruthachalam K (2012) Tomato immune receptor Ve1 recognized effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109(13):5110–5115. https://doi.org/10.1073/pnas.1119623109
Dobinson KF, Patterson NA, White GJ, Grant S (1998) DNA fingerprinting and vegetative compatibility analysis indicate multiple origins for Verticillium dahliae race 2 tomato isolates from Ontario, Canada. Mycol Res 102(9):1089–1095. https://doi.org/10.1017/S0953756297006035
Ferreira JF, Van der Merwe PC, Naude SP (1990) First report of race 2 Verticillium dahliae on tomatoes in South Africa. Plant Dis 74(7):530. https://doi.org/10.1094/PD-74-0530E
Fisher NL, Burgess LW, Toussoun TA, Nelson PE (1982) Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72(1):151–153. https://doi.org/10.1094/Phyto-72-151
Giraud T, Enjalbert J, Fournier E, Delmotte F, Dutech C (2008) Population genetics of fungal diseases of plants. Parasite 15(3):449–454. https://doi.org/10.1051/parasite/2008153449
Glass GV, Peckham PD, Sanders JR (1972) Consequences of failure to meet assumptions underlying the fixed effects analyses of variance and covariance. Rev Edu Res 42(3):237–288. https://doi.org/10.3102/00346543042003237
Gorter GJMA (1977) Index of plant pathogens and the disease they cause in cultivated plants in South Africa, vol Sci Bul. South Africa Department of Agriculture Technical Services, p 392
Grogan RG, Ioannou N, Schneider RW, Sall MA, Kimble KA (1979) Verticillium wilt on resistant tomato cultivars in California: virulence of isolates from plants and soil and relationship of inoculum density to disease incidence. Phytopathology 69(11):1176–1180. https://doi.org/10.1094/Phyto-69-1176
Hormchong N (1969) Techniques of inoculating cotton with the bacterial blight, Fusarium wilt and Verticillium wilt organisms. MSc thesis, Oklahoma State University
Iglesias-Garcia AM, Villarroel-Zeballos MI, Feng C, du Toit LJ, Correll JC (2013) Pathogenicity, virulence and vegetative compatibility grouping of Verticillium isolates from spinach seed. Plant Dis 97(11):1457–1469. https://doi.org/10.1094/PDIS-01-13-0016-RE
Inderbitzin P, Subbarao KV (2014) Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 104(6):564–574. https://doi.org/10.1094/PHYTO-11-13-0315-IA
Inderbitzin P, Bostock RM, Davis RM, Usami T, Platt HW, Subbarao KV (2011) Phylogenetics and taxonomy of the fungal vascular wilt pathogen verticillium, with the descriptions of five new species. PLoS ONE 6(12):e28341. https://doi.org/10.1371/journal.pone.0028341
Inderbitzin P, Davis RM, Bostock RM, Subbarao KV (2013) Identification and differentiation of Verticillium species and V. longisporum lineages by simplex and multiplex PCR assays. PLoS ONE 8(6):e65990. https://doi.org/10.1371/journal.pone.0065990
Jabnoun-Khiareddine H, Daami-Remadi M, Ayed F, El Mahjoub M (2007) Incidence and distribution of Verticillium dahliae races infecting tomato in Tunisia. Tunisian J of Plant Protec 2:63–70
Jiménez-Díaz RM, Mercado-Blanco J, Olivares-García C, Collado-Romero M, Bejarano-Alcázar J, Rodríguez-Jurado D et al (2006) Genetic and virulence diversity in Verticillium dahliae populations infecting artichoke in Eastern-Central Spain. Phytopathology 96(3):288–298. https://doi.org/10.1094/PHYTO-96-0288
Jiménez-Díaz RM, Cirulli M, Bubici G, Jiménez-Gasco MM, Antoniou PP, Tjamos EC (2012) Verticillium wilt, a major threat to olive production: current status and future prospects for its management. Plant Dis 96(3):304–329. https://doi.org/10.1094/PDIS-06-11-0496
Jiménez-Díaz RM, Olivares-García C, Trapero-Casas JL, Jiménez-Gasco MM, Navas-Cortés JA, Landa BB et al (2017) Variation of pathotypes and races and their correlations with clonal lineages in Verticillium dahliae. Plant Pathol 66(4):651–666. https://doi.org/10.1111/ppa.12611
Jiménez-Gasco MM, Malcolm GM, Berbegal M, Armengol J, Jiménez-Díaz RM (2014) Complex molecular relationship between vegetative compatibility groups (VCGs) in Verticillium dahliae: VCGs do not always align with clonal lineages. Phytopathology 104(6):650–659. https://doi.org/10.1094/PHYTO-07-13-0180-R
Joaquim TR, Rowe RC (1990) Reassessment of vegetative compatibility relationships among strains of Verticillium dahliae using nitrate-nonutilizing mutants. Phytopathology 80(11):1160–1166. https://doi.org/10.1094/Phyto-80-1160
Kabir Z, Bhat RG, Subbarao KV (2004) Comparison of media for recovery of Verticillium dahliae from soil. Plant Dis 88(1):49–55. https://doi.org/10.1094/PDIS.2004.88.1.49
Kim J, Park I, Lee H, Hahm Y, Yu S (2001) Identification of Verticillium dahliae and V. albo-atrum causing wilt of tomato in Korea. J of Plant Pathol17(4):222–226
Koike ST, Subbarao KV, Davis RM, Gordon TR, Hubbard JC (1994) Verticillium wilt of cauliflower in California. Plant Dis 78(11):1116–1121. https://doi.org/10.1094/PD-78-1116
Korolev N, Katan T (1997) Improved medium for selecting nitrate-non-utilizing (nit) mutants of Verticillium dahliae. Phytopathology 87(10):1067–1070. https://doi.org/10.1094/PHYTO.1997
Korolev N, Katan J, Katan T (2000) Vegetative compatibility groups of Verticillium dahliae in Israel: their distribution and association with pathogenicity. Phytopathology 90(5):529–536. https://doi.org/10.1094/PHYTO.2000.90.5.529
Lahkim Tsror L, Hazanovsky M, Mordechi-Lebiush S, Sivan S (2001) Aggressiveness of Verticillium dahliae isolates from different vegetative compatibility groups to potato and tomato. Plant Pathol 50(4):477–482. https://doi.org/10.1046/j.1365-3059.2001.00587.x
Lamprecht SC (1986) A new disease of Medicago truncatula caused by Cylindrocladium scoparium. Phytophylactica 18:111–114
Leon-Ttacca B, Arévalo-Gardini E, Bouchon A (2019) Sudden death of Theobroma cacao L. caused by Verticillium dahliae Kleb. In Peru and its in vitro biocontrol. Ciencia and Tecnología Agropecuaria 20(1):133–148. https://doi.org/10.21930/rcta.vol20_num1_art:1251
Levene H (1960) Robust tests for Equality of Variances. In: Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB (eds) Contributions to Probability and Statistics: essays in honor of Harold Hotelling. Stanford University Press, Palo Alto, pp 278–292
Ligoxigakis EK, Markakis EA (2012) Incidence and pathogenicity of races and isolates of Verticillium dahliae in Crete, southern Greece. Phytoparasitica 40:493–506. https://doi.org/10.1007/s12600-012-0250-4
Mercado-Blanco J, Rodríguez-Jurado D, Pérez-Artés E, Jiménez-Díaz RM (2001) Detection of the nondefoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR. Plant Pathol 50:609–619. https://doi.org/10.1046/j.1365-3059.2001.00601.x
Mercado-Blanco J, Rodríguez-Jurado D, Pérez-Artés E, Jiménez-Díaz RM (2002) Detection of the defoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR. Eur J Plant Pathol 108:1–13. https://doi.org/10.1023/A:1013994827836
Mercado-Blanco J, Rodríguez-Jurado D, Parrilla-Araujo S, Jiménez-Díaz RM (2003) Simultaneous detection of the defoliating and nondefoliating Verticillium dahliae pathotypes in infected olive plants by duplex, nested polymerase chain reaction. Plant Dis 87(12):1487–1494. https://doi.org/10.1094/PDIS.2003.87.12.1487
Milgroom MG, Jiménez–Gasco MM, Olivares-García C, Drott MT, Jiménez-Díaz RM (2014) Recombination between clonal lineages of the asexual fungus verticillium dahliae detected by genotyping by sequencing. PLoS ONE 9(9):e106740. https://doi.org/10.1371/journal.pone
Milgroom MG, Del Mar Jiménez-Gasco M, Olivares-García C, Jiménez-Díaz RM (2016) Clonal expansion and migration of a highly virulent, defoliating lineage of Verticillium dahliae. Phytopathology 106(9):1038–1046. https://doi.org/10.1094/PHYTO-11-15-0300-R
Nagao H, Shiraishi T, Oshima S, Koike M, Iijima T (1997) Assessment of vegetative compatibility of race-2 tomato wilt isolates of Verticillium dahliae in Japan. Mycoscience 38(4):379–385. https://doi.org/10.1007/BF02461676
O’Garro LW, Clarkson JM (1988) Pathogenicity of race-1 and race-2 tomato wilt isolates of Verticillium dahliae from different geographical origins. J Phytopathol 123(4):297–303. https://doi.org/10.1111/j.1439-0434.1988.tb04481.x
Papaioannou IA, Ligoxigakis EK, Vakalounakis DJ, Markakis EA, Typas MA (2013) Phytopathogenic, morphological, genetic and molecular characterization of a Verticillium dahliae population from Crete, Greece. Eur J Plant Pathol 136(3):577–596. https://doi.org/10.1007/s10658-013-0189-4
Qin Q-M, Vallad GE, Subbarao KV (2008) Characterization of Verticillium dahliae and V. tricorpus isolates from lettuce and artichoke. Plant Dis 92:69–77. https://doi.org/10.1094/PDIS-92-1-0069
Schnathorst WC, Mathre DE (1966) Host range and differentiation of a severe form of Verticillium albo-atrum in cotton. Phytopathology 56(10):1155–1161
Shapiro SS, Wilk MB (1965) An analysis of variance test for normality (complete samples). Biometrika 52:591–611. https://doi.org/10.2307/2333709
Short DPG, Gurung S, Maruthachalam K, Atallah ZK, Subbarao KV (2014) Verticillium dahliae race 2-specific PCR reveals a high frequency of race 2 strains in commercial spinach seed lots and delineates race structure. Phytopathology 104(7):779–785. https://doi.org/10.1094/PHYTO-09-13-0253-R
Strauss J, Labuschagne N (1995) Pathogenicity of Fusarium solani isolates on citrus roots and evaluation of different inoculum types. Toegepaste Plantwetenskap 9(2):48–52
Suaste-Dzul AP, Velosos JS, Costa H, Boiteux LS, Lourenço V, Lopes CA, Reis A (2022) Verticillium diseases of vegetable crops in Brazil: host range, microsclerotia production, molecular haplotype network, and pathogen species determination. Plant Pathol 71:1417–1430. https://doi.org/10.1111/ppa.13562
Suaste-Dzul AP, Costa H, Fonseca MEN, Boiteux LS, Reis A (2023) Mating types and physiological races of Verticillium dahliae in Solanaceae crops in Brazil. Eur J Plant Pathol 165:139–152. https://doi.org/10.1007/s10658-022-02594-8
Subbarao KV, Chassot A, Gordon TR, Hubbard JC, Bonello P, Mullin R et al (1995) Genetic relationships and cross pathogenicities of Verticillium dahliae isolates from cauliflower and other crops. Phytopathology 85(10):1105–1112. https://doi.org/10.1094/Phyto-85-1105
Tabaeizadeh Z, Agharbaoui Z, Harrak H, Poysa V (1999) Transgenic tomato plants expressing a Lycopersicon chilense chitinase gene demonstrate improved resistance to Verticillium dahliae race 2. Plant Cell Rep 19(2):197–202. https://doi.org/10.1007/s002990050733
Usami T, Momma N, Kikuchi S, Watanabe H, Hayashi A, Mizukawa M et al (2017) Race 2 of Verticillium dahliae infecting tomato in Japan can be split into two races with differential pathogenicity on resistant rootstocks. Plant Pathol 66:230–238. https://doi.org/10.1111/ppa.12576
Uys MDR (1996) The Fusarium/Verticillium root disease complex of tomato. MSc thesis, Stellenbosch University
Uys MDR, Rong IH (1995) The first report of Verticillium tricorpus as a wilt pathogen of tomato in South Africa (Abstr). SA J Science 91:xiii
Uys MDR, Rong IH, Holz G (1997) Verticillium tricorpus associated with wilt of tomato in South Africa. Afr Plant Protec 3(2):73–75
Villarroel-Zeballos MI, Feng C, Iglesias A, du Toit LJ, Correll JC (2012) Screening for resistance to Verticillium wilt in spinach and isolation of Verticillium dahliae from seed of spinach accessions. HortScience 47(9):1297–1303. https://doi.org/10.21273/HORTSCI.47.9.1297
Visser S, Hattingh MJ (1981) Relationship of inoculum of Verticillium dahliae to disease in tomato plants. Plant Soil 63(2):239–249. https://doi.org/10.1007/BF02374602
Xiao CL, Subbarao KV (1998) Relationships between Verticillium dahliae inoculum density and wilt incidence, severity, and growth of cauliflower. Phytopathology 88(10):1108–1115. https://doi.org/10.1094/PHYTO.1998.88.10.1108
Acknowledgements
This work was financed by the Agricultural Research Council, Bertie van Zyl Pty (Ltd) and the Tomato Producers’ Organisation South Africa.
Funding
Open access funding provided by Agricultural Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Ethical approval
This article is not submitted for publication elsewhere and complies with the Ethical Rules applicable for J. of plant pathology. Also, this article does not contain any studies with human participants or animals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Retief, E., Lamprecht, S. & McLeod, A. Characterisation and pathogenicity of Verticillium dahliae isolates associated with Verticillium wilt of tomato in the Limpopo Province of South Africa. J Plant Pathol 105, 1465–1481 (2023). https://doi.org/10.1007/s42161-023-01446-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-023-01446-6