Abstract
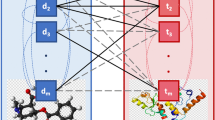
Computational drug repositioning gains more attention due to offering alternative treatments with lower development costs. This study proposes a machine learning-based drug repositioning approach by integrating various biological data. Transcriptome profiles of drug treatments and cancer patients are represented as gene expression profiles, which have been integrated into protein-protein interaction networks. The aim is to summarize molecular interaction data covered in biological networks, therefore individual networks were analyzed by attributed biased random walk (ABRW) to get low-dimensional representations of disease and drug networks. One of novel contributions this study is usage of ABRW as an embedding method in biological domain. It provides vector embeddings for drug and disease networks that have been later combined to build cancer specific drug repositioning models. We employed these supervised models to predict whether a given drug might be suggested as an alternative treatment for lung cancer or breast cancer. The experiments on new drug samples revealed more than 20 drugs as potential treatment candidates for two cancers. Comparison with another drug repositioning approach showed common predictions which have been used in some clinical trials. The application of an attributed network embedding algorithm for biological networks and its usage in the feature extraction step of a drug repositioning approach are two novel contributions of this study.

Similar content being viewed by others
References
Pan X et al (2022) Deep learning for drug repurposing: methods, databases, and applications. Wiley Interdiscip Rev Comput Mol Sci 12(4):e1597. https://doi.org/10.1002/WCMS.1597
Tanoli Z, Vähä-Koskela M, Aittokallio T (2021) Artificial intelligence, machine learning, and drug repurposing in cancer. Expert Opin Drug Discov 16(9):977–989. https://doi.org/10.1080/17460441.2021.1883585
Lotfi Shahreza M, Ghadiri N, Mousavi SR, Varshosaz J, Green JR (2018) A review of network-based approaches to drug repositioning. Br Bioinform 19(5):878–892. https://doi.org/10.1093/BIB/BBX017
Zeng X, Zhu S, Liu X, Zhou Y, Nussinov R, Cheng F (2019) DeepDR: a network-based deep learning approach to in silico drug repositioning. Bioinform 35(24):5191–5198. https://doi.org/10.1093/BIOINFORMATICS/BTZ418
Conte F, Sibilio P, Fiscon G, Paci P (2022) A transcriptome- and interactome-based analysis identifies repurposable drugs for human breast cancer subtypes. Symmetry (Basel) 14(11):2230. https://doi.org/10.3390/sym14112230
Nelson W, Zitnik M, Wang B, Leskovec J, Goldenberg A, Sharan R (2019) To embed or not: network embedding as a paradigm in computational biology. Front Genet 10:381. https://doi.org/10.3389/FGENE.2019.00381/BIBTEX
Muzio G, O’Bray L, Borgwardt K (2021) Biological network analysis with deep learning. Br Bioinform 22(2):1515–1530. https://doi.org/10.1093/BIB/BBAA257
Su XR, You ZH, Hu L, Huang YA, Wang Y, Yi HC (2021) An efficient computational model for large-scale prediction of protein-protein interactions based on accurate and scalable graph embedding. Front Genet 12:635. https://doi.org/10.3389/FGENE.2021.635451
Hamid Y, Sugumaran M (2020) A t-SNE based non linear dimension reduction for network intrusion detection. Int J Inf Technol 12(1):125–134. https://doi.org/10.1007/s41870-019-00323-9
Nerurkar P, Chandane M, Bhirud S (2022) Empirical analysis of synthetic and real networks. Int J Inf Technol 14(2):1061–1073. https://doi.org/10.1007/s41870-019-00344-4
Yue X et al (2020) Graph embedding on biomedical networks: methods, applications and evaluations. Bioinformatics 36(4):1241–1251. https://doi.org/10.1093/BIOINFORMATICS/BTZ718
Yang K, Zhao X, Waxman D, Zhao XM (2019) Predicting drug-disease associations with heterogeneous network embedding. Chaos 29(12):123109. https://doi.org/10.1063/1.5121900
Zhou R, Lu Z, Luo H, Xiang J, Zeng M, Li M (2020) NEDD: a network embedding based method for predicting drug-disease associations. BMC Bioinform 21(13):1–12. https://doi.org/10.1186/S12859-020-03682-4
Wei X, Zhang Y, Huang Y, Fang Y (2019) Predicting drug–disease associations by network embedding and biomedical data integration. Data Technol Appl 53(2):217–229. https://doi.org/10.1108/DTA-01-2019-0004
Hou C, He S, Tang K (2018) Attributed network embedding for Incomplete attributed networks. https://doi.org/10.48550/arxiv.1811.11728
Linghu B, Snitkin ES, Hu Z, Xia Y, DeLisi C (2009) Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol 10(9):1–17. https://doi.org/10.1186/GB-2009-10-9-R91
Subramanian A et al (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171(6):1437–1452. https://doi.org/10.1016/J.CELL.2017.10.049
The Cancer Genome Atlas Program-NCI (2022) https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Accessed 24 Nov 2022
Mounir M et al (2019) New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLOS Comput Biol 15(3):e1006701. https://doi.org/10.1371/JOURNAL.PCBI.1006701
Jameel R, Shobitha M, Mourya AK (2022) Predictive modeling and cognition to cardio-vascular reactivity through machine learning in indian adults with sedentary and physically active lifestyle. Int J Inf Technol 14(4):2129–2140. https://doi.org/10.1007/s41870-021-00721-y
Sharma A, Mishra PK (2022) Performance analysis of machine learning based optimized feature selection approaches for breast cancer diagnosis. Int J Inf Technol 14(4):1949–1960. https://doi.org/10.1007/s41870-021-00671-5
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. https://doi.org/10.1145/2939672
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kart, Ö., Kökçü, G., Çoçan, İ.N. et al. Application of network embedding and transcriptome data in supervised drug repositioning. Int. j. inf. tecnol. 15, 2637–2643 (2023). https://doi.org/10.1007/s41870-023-01302-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-023-01302-x