Abstract
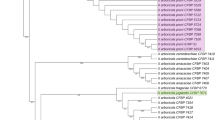
During surveys in 2018 and 2019, diseased plant materials (leaves and fruits) were collected from commercial hazelnut orchards in Samsun, Ordu, and Giresun provinces in the Black Sea region of Turkey. Bacterial isolation was performed from 188 symptomatic materials and 166 bacterial colonies with yellow color were purified on GYCA plates. Twenty-eight strains were found capable of causing lesions on bean pods, hypersensitive reaction to tobacco leaves, and disease symptoms on hazelnut seedlings. They were initially identified as X. arboricola by physiological, biochemical, and MALDI-TOF MS analyses. In PCR analysis, the strains with primers XarbQ-F/XarbQ-R and XapY17-F/XapY17-R produced a single band of 402 and 943 bp, respectively. All strains showed homogeneous band profiles for ERIC-PCR. The partial DNA sequences showed that 16 strains had more than 99% gyrB sequence similarity to strains of Xanthomonas arboricola, including the pathotype strain CFBP1159PT. The rpoD sequences were found to be highly capable of distinguishing the strains from the closely related pathovars junglandis and pruni, which showed 99.2% identity with the complete genomes of X. arboricola pv corylina strains available in Genbank. Phylogenetic tree analysis using rpoD sequences allowed separation of the hazelnut strains from other pathovars of X. arboricola. Twenty-five of the 28 strains grew on nutrient medium supplemented with 2.56 mM copper sulfate, and all were able to grow on 5 ppm streptomycin sulfate. To our knowledge, this is the most comprehensive study to characterize Turkish X. arboricola pv. corylina strains using molecular identification techniques and to determine responses to copper and streptomycin.


Similar content being viewed by others
References
Alay K, Altinyay N, Hancioglu Ö, Dündar F, Ünal A (1973) Studies on the death of the hazelnut bushes in the Black Sea region. Plant Prot Bull 13(4)
Anonymous (2016). Hazelnut cultivars. https://www.researchgate.net/profile/NerimanBeyhan/publication/322397507_Hazelnut_Cultivars/links/5a60911ba6fdcc08a43228bb/Hazelnut-Cultivars.pdf. Accessed 20 Mar 2021
Anonymous (2018a) Food and Agriculture Organization of the United Nations (FAO) FAOSTAT Database. Retrieved March 20, 2021 from https://www.fao.org/faostat/en/#data/QCL/visualize. Accessed 20 Mar 2021
Anonymous (2018b) Crop Production Statistics. Retrieved June 20, 2021 from: https://biruni.tuik.gov.tr/medas/?kn=92&locale=en. Accessed 20 June 2021
Back CG, Park MJ, Park GS, Yang CY, Park JH (2020) Complete genome sequence of Xanthomonas arboricola pv. pruni strain KACC21687 a causal agent for bacterial shot hole on peach. Korean J Microbiol 56(1):72–73
Barss HP (1915) Economic entomology. A new filbert disease in Oregon, Pamphlets.
Battilani P, Chiusa G, Arciuolo R, Somenzi M, Fontana M, Castello G, Spigolon N (2018) Diaporthe as the main cause of hazelnut defects in the Caucasus region. Phytopathol Mediterr 57(2):320–333. https://doi.org/10.14601/Phytopathol_Mediterr-22872
Behlau F, Canteros BI, Jones JB, Graham JH (2012) Copper resistance genes from different xanthomonads and citrus epiphytic bacteria confer resistance to Xanthomonas citri sub sp. citri. Eur J Plant Pathol 133(4):949–963. https://doi.org/10.1007/s10658-012-9966-8
Celenk VU, Argon, ZU, Gumus, ZP (2020) Cold pressed hazelnut (Corylus avellana) oil. In: Cold Pressed Oils Academic Press, 241–254. https://doi.org/10.1016/B978-0-12-818188-1.00020-7
Cuesta-Morrondo S, Redondo C, Palacio-Bielsa A, Garita-Cambronero J, Cubero J (2022) Complete genome sequence resources of six strains of the most virulent pathovars of Xanthomonas arboricola using long- and short- read sequencing spproaches. Phytopathology 112(8):1808–1813. https://doi.org/10.1094/PHYTO-10-21-0436-A
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S et al (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:465–469. https://doi.org/10.1093/nar/gkn180
EPPO (2004) EPPO quarantine pest ID Xanthomonas arboricola pv. corylina. Bull OEPP/EPPO Bull 34:179–181
Ertunc F, Topkaya S, Sezer A (2014) Distribution and molecular detection of apple mosaic virus in apple and hazelnut in Turkey. Afr J Biotechnol 13(31):3144–3149. https://doi.org/10.5897/AJB2013.13154
Essakhi S, Cesbron S, Fischer-Le Saux M, Bonneau S, Jacques M-A, Manceau C (2015) Phylogenetic and variable-number tandem-repeat analyses identify nonpathogenic Xanthomonas arboricola lineages lacking the canonical type III secretion system. Appl Environ Microbiol 81:5395–5410. https://doi.org/10.1128/AEM.00835-15
Fargier E, Fischer-Le Saux M, Manceau C (2011) A multilocus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer-attacking pathovars of this species. Syst Appl Microbiol 34(2):156–165. https://doi.org/10.1016/j.syapm.2010.09.001
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125(1):1–15
Fischer-Le Saux M, Bonneau S, Essakhi S, Manceau C, Jacques MA (2015) Aggressive emerging pathovars of Xanthomonas arboricola represent widespread epidemic clones distinct from poorly pathogenic strains, as revealed by multilocus sequence typing. Appl Environ Microbiol 81(14):4651–4668. https://doi.org/10.1128/AEM.00050-15
Frutos D (2010) Bacterial diseases of walnut and hazelnut and genetic resources. J Plant Pathol 92(S1):79-S85
Giovanardi D, Bonneau S, Gironde S et al (2016) Morphological and genotypic features of Xanthomonas arboricola pv. juglandis populations from walnut groves in Romagna region. Italy Eur J Plant Pathol 145:1–16. https://doi.org/10.1007/s10658-015-0809-2
Giovanardi D, Dallai D, Stefani E (2017) Population features of Xanthomonas arboricola pv. pruni from Prunus spp. orchards in northern Italy. Eur J Plant Pathol 147:761–771. https://doi.org/10.1007/s10658-016-1040-5
Gönenc S, Tanrıvermis H, Bülbül M (2006) Economic assessment of hazelnut production and the importance of supply management approaches in Turkey. J Agric Rural Dev Trop 107(1):19–32
Ivanović Ž, Popović T, Janse J, Kojić M, Stanković S, Gavrilović V, Fira D (2015) Molecular assessment of genetic diversity of Xanthomonas arboricola pv. juglandis strains from Serbia by various DNA fingerprinting techniques. Eur J Plant Pathol 141(1):133–145. https://doi.org/10.1007/s10658-014-0531-5
Jamwal S, Dutta U, Srivastava JN (2022) Major diseases of hazelnut (Corylus avellana) and their management, 1st edn. Apple Academic Press, New Jersey, pp 233–245
Kałużna M, Pulawska J, Waleron M, Sobiczewski P (2014) The genetic characterization of Xanthomonas arboricola pv. juglandis, the causal agent of walnut blight in Poland. Plant Pathol 63(6):1404–1416. https://doi.org/10.1111/ppa.12211
Kałużna M, Fischer-Le Saux M, Pothier JF, Jacques MA, Obradović A, Tavares F, Stefani E (2021) Xanthomonas arboricola pv. juglandis and pv. corylina: brothers or distant relatives? Genetic clues, epidemiology, and insights for disease management. Mol Plant Pathol 22(12):1481–1499. https://doi.org/10.1111/mpp.13073
Karahan A, Altundağ Ş, Duran H, Kılınç AO (2013) Karadeniz Bölgesinde fındık bakteriyel yanıklığı [Xanthomonas arboricola pv corylina (Miller et al.) Vauterin et al.] hastalığının yaygınlığı üzerine araştırmalar. Bitki Koruma Bülteni 53(3):159–174
Kazempour MN, Ali B, Elahinia SA (2006) First report of bacterial blight of hazelnut caused by Xanthomonas arboricola pv. corylina in Iran. J. Plant Pathol 88(3):341
Kilic, O. (1997). Economic analysis of hazelnut producing farms in lowlands of Carsamba and Terme districts of Samsun province and searching alternative feasible farm plans. Ankara University, Department of Agricultural Economics, Doctoral dissertation, Turkey.
Lamichhane JR, Varvaro L (2014) Xanthomonas arboricola disease of hazelnut: current status and future perspectives for its management. Plant Pathol 63(2):243–254. https://doi.org/10.1111/ppa.12152
Lamichhane JR, Fabi A, Ridolfi R, Varvaro L (2013) Epidemiological study of hazelnut bacterial blight in central Italy by using laboratory analysis and geostatistics. PloS ONE 8(2):e56298. https://doi.org/10.1371/journal.pone.0056298
Lee YA, Hildebrand DC, Schroth MN (1992) Use of quinate metabolism as a phenotypic property to identify members of Xanthomonas campestris DNA homology group 6. Physiol Biochem 82:971–973
Lelliot RA, Stead DE (1987) Methods for the diagnosis of bacterial diseases of plants. In: Preece TF (ed) Methods in Plant Pathology. Black Well Scientific Publications, Oxford, pp 176–177
Maes M (1993) Fast classification of plant-associated bacteria in the Xanthomonas genus. FEMS Microbiol Lett 113:161–166
Miller PW, Bollen WB, Simmons JE (1949) Filbert bacteriosis and its control (technical report 16). Agricultural Experiment Station, Oregon State College, Corvallis
Ozturk M, Soylu S (2022) A new disease of strawberry, bacterial blight caused by Erwinia amylovora in Turkey. J Plant Pathol 104:269–280. https://doi.org/10.1007/s42161-021-00994-z
Pagani MC (2004) An ABC transporter protein and molecular diagnoses of Xanthomonas arboricola pv. pruni causing bacterial spot of stone fruits. Doctoral dissertation, University of North Carolina State, Raleigh
Pavlovic M, Konrad R, Iwobi AN, Sing A, Busch U, Huber I (2012) A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol Lett 328:46–53. https://doi.org/10.1111/j.1574-6968.2011.02479.x
Pereira UP, Gouran H, Nascimento R, Adaskaveg JE, Goulart LR, Dandekar AM (2015) Complete genome sequence of Xanthomonas arboricola pv. juglandis 417, a copper-resistant strain isolated from Juglans regia L. Genome Announc 3(5):e01126-15. https://doi.org/10.1128/genomeA.01126-15
Pothier JF, Pagani MC, Pelludat C, Ritchie DF, Duffy B (2011a) A duplex PCR method for species- and pathovar-level identification and detection of the quarantine plant pathogen Xanthomonas arboricola pv. pruni. J Microbiol Methods 86:16–24. https://doi.org/10.1016/j.mimet.2011.03.019
Pothier JF, Pfluger V, Ziegler D, Tonolla M, Vogel G, Duffy B (2011b) MALDI-TOF mass spectrometry: applications for rapid bacterial identification and phylogenetic analysis. Phytopathology 101:S145
Pothier JF, Kałużna M, Prokić A, Obradović A, Rezzonico F (2022) Complete genome and plasmid sequence data of three strains of Xanthomonas arboricola pv. corylina, the bacterium responsible for bacterial blight of hazelnut. Phytopathology 112(4):956–960. https://doi.org/10.1094/PHYTO-08-21-0356-A
Prokić A, Gašić K, Ivanović MM, Kuzmznović N, Šević M, Puławska J, Obradović A (2012) Detection and identification methods and new tests as developed and used in the framework of cost873 for bacteria pathogenic to stone fruits and nuts Xanthomonas arboricola pv. corylina. J Plant Pathol 94(S1):127–133
Psallidas PG (1987) The problem of bacterial canker of hazelnut in Greece caused by Pseudomonas syringae pv. avellanae 1. EPPO Bulletin 17(2):257–261
Pulawska J, Kaluzna M, Kolodziejska A, Sobiczewski P (2010) Identification and characterization of Xanthomonas arboricola pv corylina causing bacterial blight of hazelnut: a new disease in Poland. J Plant Pathol 92(3):803–806. https://doi.org/10.4454/jpp.v92i3.331
Schaad NW, Jones JB, Chun W (2001) Laboratory guide for the identifcation of plant pathogenic bacteria, 3rd edn. American Phytopathological society (APS Press), St. Paul
Scortichini M, Rossi MP, Marchesi U (2002a) Genetic, phenotypic and pathogenic diversity of Xanthomonas arboricola pv. corylina strains question the representative nature of the type strain. Plant Pathol 51:374–381
Scortichini M, Marchesi U, Rossi MP, Di Prospero P (2002) Bacteria associated with hazelnut (Corylus avellana L.) decline are of two groups: pseudomonas avellanae and strains resembling P. syringae pv. syringae. Appl Environ Microbiol 68(2):476–484. https://doi.org/10.1128/AEM.68.2.476-484.2002b
Scortichini M, Rossi MP, Loreti S et al (2005) Pseudomonas syringae pv. coryli, the causal agent of bacterial twig dieback of Corylus avellana. Phytopathology 95:1316–1324. https://doi.org/10.1094/PHYTO-95-1316
Sezer A, Dolar FS, Lucas SJ et al (2017) First report of the recently introduced, destructive powdery mildew Erysiphe corylacearum on hazelnut in Turkey. Phytoparasitica 45:577–581. https://doi.org/10.1007/s12600-017-0610-1
Sundin GW, Bender CL (1993) Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl Environ Microbiol 59(4):1018–1024
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Vandroemme J, Cottyn B, Pothier JF, Pflüger V, Duffy B, Maes M (2013) Xanthomonas arboricola pv fragariae: what’s in a name? Plant Pathol 62(5):1123–1131. https://doi.org/10.1111/ppa.12028
Vauterin L, Hoste B, Kersters K, Swings J (1995) Reclassification of Xanthomonas. Int J Syst Evol 45(3):472–489. https://doi.org/10.1099/00207713-45-3-472
Versalovic J, Koeuth T, Lupski R (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerpriting of bacterial genomes. Nucleic Acids Res 19(24):6823–6831. https://doi.org/10.1093/nar/19.24.6823
Webber JB, Putnam M, Serdani M et al (2020) Characterization of isolates of Xanthomonas arboricola pv. corylina, the causal agent of bacterial blight, from Oregon hazelnut orchards. J Plant Pathol 102:799–812. https://doi.org/10.1007/s42161-020-00505-6
Young JM, Park DC, Shearman HM, Fargier E (2008) A multi-locus sequence analysis of the genus Xanthomonas. Syst Appl Microbiol 31:366–377. https://doi.org/10.1016/j.syapm.2008.06.004
Acknowledgements
The author thanks Prof. Dr. Soner Soylu for MALTI-TOF MS analysis of strains, Prof. Dr. Joanna Puławska for providing Polish strains L22 and 299, Prof. Dr. Aleksa Obradovic for providing Serbian strains KFB 275 and KFB 305, and Dr. A. Demirci for diligent proofreading of this manuscript.
Funding
This study was financially supported by a grant from the Office of Scientific Research Projects Coordination, Yozgat Bozok University (Project No. 6602c-ZF/18-237).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there is no conflict of interest for this submission.
Human animals rights
This article does not contain any studies with human participants or animals performed by any authors.
Informed consent
This manuscript is new and not being considered elsewhere. The authors have read and approved the submission of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Öztürk, M. Population characteristics of Xanthomonas arboricola pv. corylina strains from hazelnut orchards in Turkey. J Plant Dis Prot 130, 337–349 (2023). https://doi.org/10.1007/s41348-023-00709-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-023-00709-9