Abstract
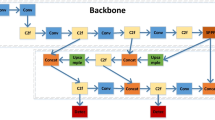
The automatic identification and diagnosis of plant disease severity are still challenging in the agricultural information system. In this study, a novel lightweight convolutional neural network (CNN)-based network with channel shuffle operation and multiple-size module (L-CSMS) is proposed for plant disease severity recognition. Specifically, the proposed stacked block consists of residual learning, channel shuffle operation, and multiple-size convolutional modules. The main contributions of this paper include the following: a lightweight and accurate network for practical plant disease severity diagnosis system is designed; it is the first attempt to incorporate the channel shuffle operation and the multiple-size convolution module into the building block as a stacked topology. Finally, the proposed lightweight CNNs-based model achieves a competitive performance over the previous works (such as ShuffleNet, MobileNet) with the accuracy of 0.906 and 0.979 on the plant disease severity dataset and PlantVillage dataset, respectively. Additionally, extensive experiments are conducted to demonstrate that the proposed method is effective for plant disease diagnosis.











Similar content being viewed by others
References
Alvarez AM (2004) Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu Rev Phytopathol 42:339–366
AlGubory KH (2014) Plant polyphenols, prenatal development and health out-comes. Biological Systems Open Access 3(1):1–2
Artzai P, Aitor AG, Maximiliam S, Amaia OB et al (2019) Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput Electron Agric 161(2019):280–290
ArunPriya C, Balasaravanan T, Thanamani AS (2012) An efficient leaf recognition algorithm for plant classification using support vector machine. In International conference on pattern recognition, informatics and medical engineering (PRIME-2012), 428–432
Barbedo JGA (2018) Factors influencing the use of deep learning for plant disease recognition. Biosys Eng 172:84–91
Bottou L (2010) Large-scale machine learning with stochastic gradient descent. Proceedings of COMPSTAT2010, 177–186
Bakhshipour A, Jafari A (2018) Evaluation of support vector machine and artificial neural networks in weed detection using shape features. Comput Electron Agric 145:153–160
Bock CH, Poole GH, Parker PE, Gottwald TR (2017) Plant disease sensitivity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit Rev Plant Sci 26:59–107
Chollet F (2016) Xception: deep learning with depthwise separable convolutions. arXiv preprint
Chaerani R, Voorrips RE (2006) Tomato early blight (alternaria solani): the pathogen, genetics, and breeding for resistance. J Gen Plant Pathol 72(6):335–347
Deng L, Yu D (2014) Deep learning: methods and applications. Foundations and Trends in Signal Processing. 3–4
Fomitcheva VW, Kühne T (2019) Beet soil-borne mosaic virus: development of virus-specific detection tools. J Plant Dis Prot 126:255–260
Mutka AM, Bart RS (2015) Image-based phenotyping of plantdisease symptoms. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00734
Gutierrez-Aguirre I, Mehle N, Delic D, Gruden K, Mumford R, Ravnikar M (2009) Real-time quantitative PCR based sensitive detection and genotype discrimination of pepino mosaic virus. J Virol Method 162:46–55
Gwo CY, Wei CH (2013) Plant identification through images: using feature extraction of key points on leaf contours. Appl Plant Sci 1(11):1200005
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE Conf Comput Vision Pattern Recognit. 770–778
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang WJ, Weyand T, Andreetto M, Adam H (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint
Huang G, Liu S, van der Maaten L, Weinberger KQ (2017) Condensenet: an efficient densenet using learned group convolutions. arXiv preprint
Huang G, Liu Z, Weinberger KQ, van der Maaten L (2017) Densely connected convolutional networks. IEEE Conf Comput Vision Pattern Recognit. 2261–2269
Hu J, Shen L, Sun G (2017) Squeeze-and-excitation networks. arXiv preprint
Harvey CA, Rakotobe ZL, Rao NS, Dave R, Mackinnon JL (2014) Extreme vulnerability of smallholder farmers to agricultural risks and climate change in Madagascar. Philos Trans R Soc Lond 369(1639):20130089
Johannes A, Picon A, Alvarez-Gila A, Echazarra J et al (2017) Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput Electron Agric 138:200–209
Jayme GAB (2019) Plant disease identification from individual lesions and spots using deep learning. Biosys Eng 180:96–107
Ketkar N (2017) Introduction to PyTorch. Deep Learning with Python. Apress, Berkeley, CA, pp 195–208
Krizhevsky A, Sutskever I, Hinton G (2012) ImageNet classification with deep convolutional neural networks. In: International Conference on Neural Information Processing Systems Curran Associates Inc. 1097–1105
Krishna S, Indra G, Sangeeta G (2010) SVM-BDT PNN and fourier moment technique for classification of leaf shape. Int J Signal Process, Image Process, Pattern Recognit 3(4):341–348
Kumar N, Belhumeur PN, Biswas A, Jacobs DW, et al. (2012) Leafsnap: a computer vision system for automatic plant species identification
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nat 521:436–444
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Liang QK, Xiang S, Hu YC, Coppola G, Zhang D, Sun W (2019) PD2SE-Net: computer-assisted plant disease diagnosis and severity estimation network. Comput Electron Agric 157:518–529
Long J, Shelhamer E, Darrell T (2014) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651
Lu Y, Yi S, Zeng NY, Liu YR, Zhang Y (2017) Identification of rice diseases using deep convolutional neural networks. Neurocomput 267:378–384
Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, Villa P, Stroppiana D, Boschetti M, Goulart LR, Davis CE, Dandekar AM (2014) Advanced methods of plant disease detection. Agron Sustain Dev 35(1):1–25
Mallah C, Cope J, Orwell J (2013) Plant leaf classification using probabilistic integration of shape, texture and margin features. Pattern Recognit, Appl., p 3842
Ma N, Zhang X, Zheng HT, Sun J (2018) Shufflenet v2: Practical Guidelines for Efficient CNN Architecture Design. arXiv preprint arXiv: 1807 11164
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1–7
Oerke EC (2006) Crop losses to pests. J Agric Sci 144:31–43
Owomugisha G, Mwebaze E (2016) Machine Learning for Plant Disease Incidence and Severity Measurements from Leaf Images. Proc 2016 15th IEEE Int Conf Mach Learn Appl (ICMLA), Anaheim, CA, USA, 18–20
Puja D, Saraswat M, Arya K (2012) Automatic agricultural leaves recognition system. Adv Intell Sys Comput 201:123–131
Ragupathy S, Herbarium OAC (2016) Biodiversity institute of ontario from university of Guelph
Ren S, He K, Girshick R, Sun J (2017) Faster r-cnn: towards real-time object detection with region proposal networks. IEEE Trans Patt Anal Mach Intell 39(6):1137–1149
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang ZH, Karpathy A, Khosla A, Bernstein M, Berg AC, Li FF (2015) Imagenet large scale visual recognition challenge. Int J Comput Vision 115(3):211–252
Saleem G, Akhtar M, Ahmed N, Qureshi WS (2019) Automated analysis of visual leaf shape features for plant classification. Comput Electron Agric 157:270–280
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. IEEE Conf Comput Vision Pattern Recognit. 1–9
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2016) Inception-v4, Inception-ResNet and the impact of residual connections on learning. Proc of the Thirty-First AAAI Conf Artif Intell. 4278–4284
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L (2018) Inverted Residuals and Linear Bottlenecks: Mobile Networks for Classification, Detection and Segmentation. arXiv preprint
Sa I, Ge Z, Dayoub F, Upcroft B, Perez T, McCool C (2016) Deepfruits : a fruit detection system using deep neural networks. Sens 16(8):1222
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci 2016:1–11
Silva PFB, Marcal ARS, da Silva MRA (2013) Evaluation of features for leaf discrimination. Int Conf Image Anal Recognit 2013:197–204
Selvaraju RR, Cogswell M, Das A, Vedantam R, et al. (2016) Grad-cam: visual explanations from deep networks via gradient-based localization. arXiv preprint
Wu J, Zhou J, Jiao Z, Fu J, Guo F (2019) Identification of the pathogens causing konjac dry rot in China. J Plant Dis Prot 126:263–264
Wang H, Li G, Ma Z, Li X (2012) Application of neural networks to image recognition of plant diseases. Int Conf Syst Inform. 2012(2159):2164
Wang G, Sun Y, Wang J (2017) Automatic image-based plant disease severity estimation using deep learning. Comput. Intell, Neurosci, p 2917536
Xie S, Girshick R, Dolla ́r P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. IEEE Conf Comput Vision and Pattern Recognit 1492–1500
Zhang X, Zhou X, Lin M, Sun J (2017) Shufflenet: An extremely efficient convolutional neural network for mobile devices. arXiv preprint
Funding
This work was supported in part by the National Nature Science Foundation of China (NSFC 62073129, 61673163), the Chang-Zhu-Tan National Indigenous Innovation Demonstration Zone Project (2017XK2102), and the Nature Science Research Project of Anhui Province (1808085QF195).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Rights and permissions
About this article
Cite this article
Xiang, S., Liang, Q., Sun, W. et al. L-CSMS: novel lightweight network for plant disease severity recognition. J Plant Dis Prot 128, 557–569 (2021). https://doi.org/10.1007/s41348-020-00423-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-020-00423-w