Abstract
In neuroscience, clustering subjects based on brain dysfunctions is a promising avenue to subtype mental disorders as it may enhance the development of a brain-based categorization system for mental disorders that transcends and is biologically more valid than current symptom-based categorization systems. As changes in functional connectivity (FC) patterns have been demonstrated to be associated with various mental disorders, one appealing approach in this regard is to cluster patients based on similarities and differences in FC patterns. To this end, researchers collect three-way fMRI data measuring neural activation over time for different patients at several brain locations and apply Independent Component Analysis (ICA) to extract FC patterns from the data. However, due to the three-way nature and huge size of fMRI data, classical (two-way) clustering methods are inadequate to cluster patients based on these FC patterns. Therefore, a two-step procedure is proposed where, first, ICA is applied to each patient’s fMRI data and, next, a clustering algorithm is used to cluster the patients into homogeneous groups in terms of FC patterns. As some clustering methods used operate on similarity data, the modified RV-coefficient is adopted to compute the similarity between patient specific FC patterns. An extensive simulation study demonstrated that performing ICA before clustering enhances the cluster recovery and that hierarchical clustering using Ward’s method outperforms complete linkage hierarchical clustering, Affinity Propagation and Partitioning Around Medoids. Moreover, the proposed two-step procedure appears to recover the underlying clustering better than (1) a two-step procedure that combines PCA with clustering and (2) Clusterwise SCA-ECP, which performs PCA and clustering in a simultaneous fashion. Additionally, the good performance of the proposed two-step procedure using ICA and Ward’s hierarchical clustering is illustrated in an empirical fMRI data set regarding dementia patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Nowadays, several research questions in neuroscientific studies call for a clustering of subjects based on high-dimensional—big—brain data. For example, a promising trend in clinical neuropsychology is to categorize (and subtype) mental disorders based on brain dysfunctions instead of on symptom profiles (only) (see Sect. 2.1). Relevant brain dysfunctions in this regard are the changes in functional connectivity (FC), where FC refers to the synchronized activity over time of spatially distributed brain regions (Barkhof et al. 2014). These changes in FC have been demonstrated (see Sect. 2.1) to be related to various neuropsychiatric diseases—and subtypes therein—such as depression and dementia (Greicius et al. 2004). As such, these FC pattern changes can be used to cluster patients and to identify the corresponding distinct mental disorder categories and subtypes. In particular, each obtained patient cluster may represent a distinct mental disorder/subtype. Moreover, using an unsupervised clustering technique that ignores the existing patient labels, the obtained categories and subtypes may differ from the symptom-based categories and subtypes and may account for the large heterogeneity in symptoms, disease courses and treatment responses encountered within the existing categories and subtypes (e.g., Alzheimer’s disease and frontotemporal dementia).
Key to the clustering is the identification of relevant FC pattern changes. To capture these FC patterns, researchers often collect functional Magnetic Resonance Imaging (fMRI) data for multiple patients. Such data, which can be arranged in a three-way array (see Fig. 1), represent Blood Oxygen Level Dependent (BOLD) signal changes for a large number of brain locations (voxels) that are measured over time for multiple patients at rest or while they are performing a particular task. A commonly used method to extract FC patterns from fMRI data is Independent Component Analysis (ICA), which reduces the data to a smaller set of independent components which represent brain regions that show synchronized activity (Mckeown et al. 1998; Beckmann and Smith 2004; Beckmann 2012). ICA has successfully been applied in resting-state fMRI studies to both investigate cognition and abnormal brain functioning (Smith et al. 2009; Filippini et al. 2009) and to disentangle noise (e.g., machine artefacts, head motion, physiological pulsation and haemodynamic changes induced by different processes) from relevant signal (Calhoun et al. 2001, 2009; Erhardt et al. 2011).
Three-way data representation of a multi-subject fMRI data set. The rows (first way) represent small areas in the brain known as voxels (\(v = 1, \dots , V\)). The columns (second way) represent time points (\(t = 1, \dots , T\)). The slices (third way) refer to subjects/patients (\(i = 1, \dots , N\))
A new way of categorizing/subtyping mental disorders consists of collecting fMRI data from a set of patients and clustering these patients into clusters that are homogeneous with respect to the FC patterns underlying the data of the patients. In particular, patients with similar FC patterns should be clustered together, whereas patients exhibiting patterns that are qualitatively different should be allocated to different clusters. Such an approach clearly differs from the existing approaches for clustering fMRI data in that the existing approaches only focus on clustering voxels or brain regions (Mezer et al. 2009) or on clustering functional networks (Esposito et al. 2005) but do not allow to cluster patients in terms of FC patterns. Existing methods that allow for a patient clustering use clinical symptom information (van Loo et al. 2012) or measures derived from fMRI data (i.e., graph-theoretical measures, like path length and centrality), but not raw fMRI data, as input, and, as a consequence, do not focus on underlying FC patterns.
Due to the three-way nature of multi-subject fMRI data, classical clustering methods such as k-means (Hartigan and Wong 1979), hierarchical clustering (Sokal and Michener 1958) and model-based clustering (Fraley and Raftery 2002; Banfield and Raftery 1993; McLachlan and Basford 1988) are not suitable for clustering patients based on FC patterns underlying fMRI data since these methods require two-way data (e.g., patients measured on a set of variables or (dis)similarities between patient pairs) as input. Converting three-way data to two-way data, a procedure called matricizing (Kiers 2000), is not a panacea as the classical clustering methods have large difficulties dealing with the large number of ‘variables’ created in that way (i.e., the number of voxels times the number of time points, which easily can exceed millions). Moreover, matricizing multi-subject fMRI data implies the loss of spatio-temporal information that is relevant for the clustering of the patients. As such, a proper method for clustering three-way data is needed (see Sect. 2.2 for a discussion of existing three-way clustering methods that are not appropriate for the task at hand).
In the current study, therefore, we propose a two-step procedure where, first, FC patterns are extracted by performing ICA on the data of each patient separately, and, next, the patients are clustered based on similarities and dissimilarities between the patient-specific FC patterns. To determine the degree of (dis)similarity between—the estimated FC patterns of—two patients, the modified RV-coefficient is used, which is a matrix correlation quantifying the linear relation between matrices—instead of variables—that shows favourable properties for high-dimensional data (Smilde et al. 2009). For the clustering, Affinity Propagation (AP; Frey and Dueck 2008) is used, which is a relatively new clustering method that has been shown to outperform the popular k-means algorithm for clustering brain functional activation (Zhang et al. 2011), genetic data and images/faces (Frey and Dueck 2008). A related method that also combines clustering with ICA is mixture ICA (Lee et al. 1999). This method, however, differs from our proposed two-step procedure in two ways: (1) mixture ICA can only be used for single-subject fMRI data, and (2) for the clustering it uses a mixture analysis (i.e., model-based) approach instead of a k-means-like approach (for a discussion and comparison of both types of clustering methods, see Steinley and Brusco 2011). Another related method is group ICA (Calhoun et al. 2001, 2009) in which the data of a (known) group of patients are concatenated and afterwards subjected to ICA. This method is not appropriate for the task at hand as it assumes that the patient clusters (e.g., Alzheimer vs. frontotemporal dementia patients) are known beforehand.
The goal of this paper is to evaluate in an extensive simulation study and in an illustrative application to fMRI data regarding dementia patients the performance of our two-step procedure using AP and to compare AP to commonly used methods from the family of hierarchical clustering and k-means type of clustering methods. Moreover, to demonstrate that the ICA decomposition step is a vital step for uncovering the true clusters in terms of the FC patterns underlying fMRI data, the proposed two-step procedure is compared to a procedure in which the fMRI data are clustered (1) without performing ICA, and (2) after reduction with PCA (instead of ICA). The two-step procedure is also compared to Clusterwise SCA, which performs clustering and PCA data reduction simultaneously (for a description of this method, see Sect. 2.2).
The remainder of this paper is organized as follows: in the next section, some background on brain dysfunctions and their potential for categorizing mental disorders is discussed (Sect. 2.1) and existing clustering methods for three-way data are sketched (Sect. 2.2). Next, in Sect. 3, ICA for analysing fMRI data of a single patient is discussed, together with the modified RV-coefficient for computing the (dis)similarity between FC patterns. Further, the four clustering methods that are used in this study are briefly described: (1) Affinity Propagation (AP; Frey and Dueck 2008), (2) Partitioning Around Medoids (PAM; Kaufman and Rousseeuw 1990), (3) hierarchical clustering using Ward’s method (Ward 1963) and (4) complete linkage hierarchical clustering. In the fourth section, the performance of the proposed two-step procedure is evaluated by means of an extensive simulation study. Next, in Sect. 5, the proposed two-step procedure is illustrated on empirical fMRI data regarding dementia patients. Finally, implications and limitations of the proposed procedure are discussed, along with directions for further research.
2 Background
2.1 Brain dysfunctions as the basis for categorizing mental disorders
Until recently, mainly symptom information has been used to categorize mental disorders (e.g., DSM-V; American Psychiatric Association 2013). Many psychiatric and neurocognitive disorders (e.g., depression, schizophrenia and dementia), however, show a large variability in symptoms, disease courses and treatment responses. This substantial clinical heterogeneity, which is caused by the weak links that exist between the current diagnostic categories and the underlying biology of mental disorders (see, for example, Craddock et al. 2005; Happé et al. 2006), questions the validity of the current symptom-based diagnostic categorization systems for mental disorders. As brain dysfunctions have been found to be important predisposing/vulnerability factors for many psychiatric disorders (Marín 2012; Millan et al. 2012), a way to obtain a biologically more valid diagnostic system is to base the categorization on similarities and differences between patients in brain (dys)functioning. This shift from symptom- to brain-based categorization is a crucial prerequisite for and connects well with the emerging trend of personalized psychiatry, also called precision psychiatry (Fernandes et al. 2017). Note that this shift clearly links up with recent modern mental health initiatives, such as the National Institute of Mental Health’s Research Domain Criteria in psychiatry (RDoC; Insel et al. 2010; Cuthbert 2014), the Precision Medicine Initiative (Collins and Varmus 2015) and the European Roadmap for Mental Health Research (ROAMER; Schumann et al. 2014). Additionally, as brain dysfunctions occur at pre-symptomatic stages (i.e., before structural and cognitive changes become apparent) for most mental disorders (Marín 2012; Damoiseaux et al. 2012; Drzezga et al. 2011) and are predictive for treatment response (Liston et al. 2014; Downar et al. 2014; McGrath et al. 2013), disposing of brain-based diagnostic categories allows for the early detection of subjects at risk for a particular disorder and may advance evidence-based treatments and outcomes for patients.
Using brain dysfunctions as the basis for a categorization of mental disorders is promising as recent scientific studies provided ample evidence for the relation between mental disorders and brain dysfunctions. Especially relevant in this regard are changes in FC patterns, which have been showed to be prevalent in many mental disorders (for an overview, Seeley et al. 2009; Greicius 2008; Zhang and Raichle 2010; Deco and Kringelbach 2014), like schizophrenia (Lynall et al. 2010; Jafri et al. 2008), panic disorder and social anxiety disorder (Veer et al. 2011; Pannekoek et al. 2013), Alzheimer’s disease and dementia (Pievani et al. 2011; Greicius et al. 2004; Rombouts et al. 2005), major depression (Kaiser et al. 2015; Veer et al. 2010; Miller et al. 2015) and autism spectrum disorder (Lee et al. 2017; Weng et al. 2010). Moreover, recently, Drysdale et al. (2017) and Tokuda et al. (2018) demonstrated that, using (mainly) information on (dys)functional connectivity patterns, neurophysiological subtypes of depression—called biotypes—could be derived that transcend current diagnostic symptom-based depression (sub)categories. These biotypes, which cannot be detected using symptom information only, are robust over time, are related to differential clinical symptom profiles and are predictive for treatment response. Taking this evidence together, a promising avenue for advancing precision psychiatry is to construct a brain-based categorization system for mental disorders, which may complement the existing consensus on symptom-based categorizations. To this end, patients should be clustered based on similarities and differences in their underlying FC patterns such that each patient cluster (hopefully) represents a specific mental disorder category or subtype thereof. This is a novel strategy for clustering patients as current approaches in this regard—except for the two studies mentioned earlier—mainly resort to clinical symptom profiles (for example, see, van Loo et al. 2012; Schacht et al. 2014) instead of to brain dysfunctions.
2.2 Methods for clustering three-way data
To cluster three-way data, Kroonenberg (2008) and Viroli (2011) proposed a model-based procedure. However, both these procedures cannot handle the large number of voxels—50,000 or more—typically measured in fMRI data. Moreover, these methods assume some form of multivariate normality, which is often not realistic for fMRI data. Another method that enables the clustering of three-way data is Clusterwise Simultaneous Component Analysis (for example, Clusterwise SCA-ECP; De Roover et al. 2012). This method combines SCA and clustering in such a way that a data reduction in two modes (e.g., voxels and time) and a clustering along the third mode (e.g., patients) are achieved simultaneously. Although Clusterwise SCA may result in a useful clustering of the patients, the associated components that are simultaneously estimated by this method, in general, will not yield a good representation of the FC patterns underlying fMRI data. This is caused by the fact that these models do not seek for components that are non-Gaussian and independent in the spatial domain, which is an attractive feature of (spatial) ICA. Note that FC patterns that are related to mental disorders often are non-Gaussian and show independence in the spatial domain.
3 Methods
3.1 Independent component analysis (ICA) of a single subject’s data
ICA (in particular, spatial ICA) decomposes a multivariate signal into statistically independent components and their associated time courses of activation (see Fig. 2). A critical assumption of the ICA model is that the components underlying the data follow a non-Gaussian distribution and are statistically independent of each other. As such, ICA is able to separate systematic information in the data from irrelevant sources of variability such as noise. For fMRI data, the systematic information represents functionally connected brain regions (FC patterns) that are independent in the spatial domain (i.e., spatial ICA) -also called spatial maps- and their corresponding time courses.
The ICA model for a single patient. A multivariate measured signal \({\mathbf {X}}\) (the BOLD response measured over time for a set of voxels) is assumed to result from a linear mixing of an underlying source signal matrix \({\mathbf {S}}\) with a mixing matrix \({\mathbf {A}}\). The components of \({\mathbf {S}}\) refer to underlying FC patterns whereas the components of \({\mathbf {A}}\) represent the associated underlying fMRI time courses of activation
More rigorously defined, ICA is a multivariate technique that aims at finding a linear representation of the data such that the statistical dependency between non-normally distributed components is minimized (Jutten and Herault 1991; Comon 1994). In the general ICA model for fMRI data of a single patient (with V voxels and T volumes), the observed signal mixture \({\mathbf {X}}\)\((V \times T)\) is assumed to be the result of a linear mixing of Q independent (non-Gaussian) source signals \({\mathbf {S}}\,(V \times Q)\) by means of a mixing matrix \({\mathbf {A}}\,(T \times Q)\). The general ICA model is defined as (with \((\mathbf {\cdot })^T\) denoting the matrix transpose)
The unknown source signals \({\mathbf {S}}\) can be computed by multiplying the observed mixed signals \({\mathbf {X}}\) with a weight matrix \({\mathbf {W}}\), which equals the pseudo-inverse (Golub and Van Loan 2012) of \({\mathbf {A}}\) (with the pseudo-inverse of a matrix being indicated by \(^\dagger \)):
Several methods have been proposed to estimate the weight matrix \({\mathbf {W}}\) (and hence \({\mathbf {S}}\)). The idea behind these methods is to make each column of \(\mathbf {XW}^T\) as non-Gaussian as possible, which -due to the Central Limit Theorem- results in the columns of \(\mathbf {XW}^T\) containing the independent components underlying the data (for a more theoretical explanation, see Hyvärinen and Oja 2000, Sect. 4.1). The proposed methods differ in the criterion that they use to measure the extent to which a distribution is non-Gaussian. One of the most popular and fastest methods is known as fastICA (Hyvärinen 1999). This method uses negentropy as a measure for non-Gaussianity. The negentropy (also denoted as negative entropy) of a distribution, indeed, measures the “distance” of a distribution to the normal distribution, with larger values indicating a distribution that is “further away” from the Gaussian distribution. Note that a normally distributed (Gaussian) random variable has a negentropy, which is always nonnegative, of zero; hence, maximizing the negentropy of (the columns of) \(\mathbf {XW}^T\) ensures that the estimated components (in \(\mathbf {XW}^T\)) are as non-Gaussian, and, as a result, as independent from each other as possible. FastICA achieves this by means of a fast fixed-point algorithm (Hyvärinen 1999). As a pre-processing step, the observed mixture \({\mathbf {X}}\) is often centered and pre-whitened (i.e., decorrelation and scaling to unit variance), which results in a serious decrease in the computational complexity of the estimation procedure to find the independent components (Hyvärinen et al. 2001; Beckmann and Smith 2004). Indeed, after pre-whitening, the vectors of \(\mathbf {XW}^T\) are ensured to be mutually orthogonal and of unit variance and the independent components can be identified by estimating a rotation matrix that maximizes the negentropy of \(\mathbf {XW}^T\). Note that in contrast to PCA, ICA uses higher order information (i.e, third and higher moments of the data) to estimate the components and, as a consequence, does not have rotational freedom of the components. Similar as in PCA, when performing ICA, one has to decide upon the number of components Q to extract. Most popular methods in this regard are based on searching for an elbow in a scree plot that displays the ordered eigenvalues—estimated from the data covariance matrix—against their rank number. Other popular methods are based on applying a Bayesian model selection procedure (Beckmann and Smith 2004) within the context of Probabilistic Principal Component Analysis (PPCA; Tipping and Bishop 1999) or on information theoretic measures, such as AIC (see, for example, Li et al. 2007).
3.2 Computing a (dis)similarity matrix with the modified-RV coefficient
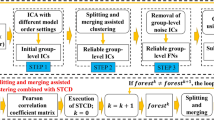
After performing ICA —with the same number of components Q— to the data \({\mathbf {X}}_i\) of each patient i\((i = 1 \dots N)\), which results in estimated patient specific FC patterns \(\hat{{\mathbf {S}}}_i^\text {ICA}\) and mixing matrices \(\hat{{\mathbf {A}}}_i^\text {ICA}\) (see Fig. 2), clustering is performed on the estimated FC patterns contained in the \(\hat{{\mathbf {S}}}_i^\text {ICA}\)’s. Similarly, one could choose to first perform PCA (with the same Q) on each data matrix \({\mathbf {X}}_i\) such that a component score matrix \(\hat{{\mathbf {S}}}_i^\text {PCA}\) and a loading matrix \(\hat{{\mathbf {A}}}_i^\text {PCA}\) is estimated for each patient. Subsequently, a clustering method is performed on the FC patterns contained in the component score matrices. As several clustering methods, such as Affinity Propagation (see later), need a similarity matrix \({\mathbf {D}}\) as input (see Fig. 3), for each pair of patients, the modified RV-coefficient (Smilde et al. 2009) is computed between the estimated \(\hat{{\mathbf {S}}}_i\)’s of both pair members (see lower panel of Fig. 3). Next, the modified RV-values for all patient pairs (i, j) \((i, j = 1 \dots N)\) are stored in the \(N \times N\) similarity matrix \({\mathbf {D}}\). The modified RV-coefficient is a matrix correlation that provides a measure of similarity of matrices.Footnote 1 The modified RV ranges between \(-1\) and 1, with 1 indicating perfect agreement and 0 agreement at chance level.Footnote 2 As the hierarchical clustering and Partitioning Around Medoids clustering methods need a dissimilarity matrix \({\mathbf {D}}^{*}\) -instead of a similarity matrix- as input, each similarity value \(d_{ij}\) of \({\mathbf {D}}\) is converted into a dissimilarity value by subtracting the value of the similarity value from 1 (i.e., \(d^{*}_{ij} = 1 - d_{ij}\)).
Schematic overview of the two-step clustering procedure. A similarity matrix \({\mathbf {D}}\) is constructed by computing all (patient) pairwise modified-RV coefficients between either the full data \({\mathbf {X}}_i\) (upper panel) or the ICA or PCA reduced component score matrices \(\hat{{\mathbf {S}}}_i\) (lower panel). In particular, each entry \(d_{ij}\) of \({\mathbf {D}}\) contains the value of the RV coefficient between \({\mathbf {X}}_i\) and \({\mathbf {X}}_j\) (\(\hat{{\mathbf {S}}}_i\) and \(\hat{{\mathbf {S}}}_j\)), with i and j referring to two patients (\(i,j = 1, \dots , N\)). Subsequently, a partitioning of the patients is obtained by applying a clustering method to either \({\mathbf {D}}\) (for AP) or \({\mathbf {D}}^*\) (for PAM, HCW and HCC), with the elements \(d^{*}_{ij}\) of dissimilarity matrix \({\mathbf {D}}^*\) being \(d^{*}_{ij} = 1 - d_{ij}\)
To determine whether performing dimension reduction through ICA/PCA before clustering the patients outperforms a strategy in which the patients are clustered based on the original fMRI data directly, also a similarity matrix \({\mathbf {D}}\) (and \({\mathbf {D}}^{*}\)) is constructed with entries being the modified RV-coefficients between the original \({\mathbf {X}}_i\)’s (see upper panel of Fig. 3).
3.3 Clustering methods
After computing \({\mathbf {D}}\) and \({\mathbf {D}}^{*}\), the patients are clustered into homogeneous groups by means of the following three clustering methods: Hierarchical Clustering (HC), Partitioning Around Medoids (PAM) and Affinity Propagation (AP). HC and PAM—which is closely related to k-means—were selected because they can be considered as standard methods that are commonly used. AP was included in the study because it is considered as a ‘new’ clustering method that has great potential for psychological and neuroscience data (Frey and Dueck 2008; Li et al. 2009; Santana et al. 2013). Model-based clustering methods were not included because they are less appropriate to deal with the format of the data (i.e., a high-dimensional data matrix instead of a vector per patient) and cannot handle a (dis)similarity matrix as input.
Agglomerative Hierarchical Clustering. Using a dissimilarity matrix as input (e.g., Euclidean distances between objects), agglomerative hierarchical clustering produces a series of nested partitions of the data by successively merging objects/object clusters into (larger) clusters.Footnote 3 Here, each object starts as a single cluster and at each iteration the most similar pair of objects/object clusters are merged together into a new cluster (Everitt et al. 2011). To determine the dissimilarity between object clusters, agglomerative clustering uses various linkage functions (for a short overview see, Everitt et al. 2011). For the current study, complete linkage and Ward’s method (Ward 1963) are used. The former method defines the dissimilarity between clusters A and B as the maximal dissimilarity \(d_{ij}\) encountered among all pairs (i, j), where i is an object from cluster A and j an object from cluster B (Kaufman and Rousseeuw 1990). In Ward’s method, object clusters are merged in such a way that the total error sum of squares (ESS) is minimized at each step (Everitt et al. 2011). Here, the ESS of a cluster is defined as the sum of squared Euclidean distances between objects i of that cluster and the cluster centroid (Murtagh and Legendre 2014). For hierarchical clustering, the number of clusters K has to be specified and methods known as stopping rules have been specially developed to this end (Mojena 1977).
Partitioning Around Medoids. PAM aims at partitioning a set of objects into K homogeneous clusters (Kaufman and Rousseeuw 1990). PAM identifies K objects in the data—called medoids- that are centrally located in—and representative for—the clusters. To this end, the average dissimilarity between objects belonging to the same cluster is minimized. The PAM method yields several advantages over other well-known partitioning methods such as k-means (Hartigan and Wong 1979) and is, therefore, included in this study instead of k-means. First, the medoids obtained by PAM are more robust against outliers than the centroids resulting from k-means (van der Laan et al. 2003). Second, in PAM the ‘cluster centers’ or medoids are actual objects in the data, whereas in k-means the centroids do not necessarily refer to actual data objects. From a substantive point of view, actual data objects as cluster representatives are preferred over centroids that are often hard to interpret. Finally, PAM can be used with any specified distance or dissimilarity metric, such as Euclidean distance, correlations and -relevant to the current study- modified RV-coefficients. The k-means method, however, is restricted to the Euclidean distance metric, which is problematic especially for high-dimensional data such as fMRI data. Indeed, as shown in Beyer et al. (1999), when the dimensionality of the data increases, the (Euclidean) distance between nearby objects approaches the distance between objects ‘further away’, implying all objects becoming equidistant from each other. Regarding selecting the optimal number of clusters K, several methods have been developed. A well-known method, for example, is the Silhouette method that determines how well an object belongs to its allocated cluster—expressed by the silhouette width \(s_{i}\)—compared to how well it belongs to the other clusters (Rousseeuw 1987). An optimal number of K can be determined by taking the K from the cluster solution that yields the largest average silhouette width (i.e., the average of all \(s_i\)’s of a clustering).
Affinity Propagation. A relatively new clustering method is known as Affinity Propagation (AP; Frey and Dueck 2008) and it is similar to other clustering methods such as p-median clustering (Köhn et al. 2010; Brusco and Steinley 2015). AP takes as input (1) a set of pairwise similarities between data objects and (2) a set of user-specified values for the ‘preference’ of each object (see further). AP aims at determining so called ‘exemplars’ which are actual data objects, with each exemplar being the most representative member of a particular cluster. AP considers all available objects as potential exemplars and, in an iterative fashion, gradually selects suitable exemplars by exchanging messages between the data objects (Frey and Dueck 2008). At the same time, for the remaining (non-exemplar) objects, the algorithm gradually determines the cluster memberships. More precisely, the objects are considered to be nodes in a network and two types of messages are exchanged between each pair of nodes (i,j) in the network: messages regarding (1) the responsibility which reflects the appropriateness of a certain object j to serve as an exemplar for object i, and (2) the availability which points at the properness for object i to select object j as its exemplar. At each iteration of the algorithm, the information in the messages (i.e., the responsibility and the availability of an object in relation to another object) is updated for all object pairs. As such, at each iteration, it becomes more clear which objects can function as exemplars and to which cluster each remaining object belongs. The procedure stops after a fixed set of iterations or when the information in the two types of messages is not updated anymore for some number of iterations (for a more technical description of the method, readers can consult Frey and Dueck 2008). A nice feature of AP is that it simultaneously estimates the clusters as well as the number of clusters. However, it should be noted that the number of clusters that is obtained by the algorithm depends on the user-specified values for the ‘preference’ of each object. This preference value determines how likely a certain object is chosen as a cluster exemplar, with higher (lower) values implying objects being more (less) likely to function as a cluster exemplar. As such, giving low (high) preference values to all objects will result in a low (high) number of clusters K. Note that implementations of AP exist in which the user can specify the desired K prior to the analysis (Bodenhofer et al. 2011). In these implementations, a search algorithm determines the optimal preference values such that the final number of clusters K equals the user specified number of clusters K (i.e., the final K can be decreased/increased by choosing lower/higher preference values).
4 Simulation study
4.1 Problem
In this section, a simulation study is presented in which it is investigated whether and to which extent the aforementioned two-step clustering procedure is able to correctly retrieve the ‘true’ cluster structure from the data. It is also evaluated whether or not using ICA for data reduction prior to the clustering enhances the recovery of the true cluster structure. Herewith, also PCA and Clusterwise SCA, which are related techniques for data reduction, are investigated. Furthermore, it is studied whether the recovery performance of the two-step procedure depends on the following data characteristics: (1) the number of clusters, (2) equal or unequal cluster sizes, (3) the degree to which clusters overlap, and (4) the amount of noise present in the data.
Based on previous research, it can be expected that the recovery of the true cluster structure deteriorates when there are a larger number of clusters underlying the data and/or when the data are noisier (Wilderjans et al. 2008; De Roover et al. 2012; Wilderjans et al. 2012; Wilderjans and Ceulemans 2013). Moreover, with respect to the size of the clusters, it can be expected that a better recovery will be obtained when the clusters are of equal size and this especially for the hierarchical clustering method using Ward’s method (Milligan et al. 1983). Further, when the true clusters show more overlap, all clustering methods are expected to deteriorate since more overlap makes it harder to find and separate the true clusters (Wilderjans et al. 2012, 2013). Finally, reducing the data with ICA before clustering is expected to outperform Clusterwise SCA and reducing the data with PCA prior to clustering as ICA better captures the FC patterns underlying the data, which are independent and Non-Gaussian, than PCA-based methods.
As mentioned before, when performing ICA (or PCA), one has to decide on the optimal number of independent components Q to extract. Often, a procedure for estimating the optimal number of components Q, like the scree plot (see earlier), is used to this end. To investigate the effect of under- and overestimating the true number of components \(Q^\text {true}\) (which was kept equal across patients) on the true clustering, we re-analysed a small portion of the simulated data sets and varied the number of components Q used for ICA. For this simulation study, it can be expected that the underlying cluster structure is recovered to a large extent when the selected number of components Q equals or perhaps is close to the true number of components \(Q^\text {true}\). In other cases, however, it is expected that the true clusters are not retrieved well.
4.2 Design and procedure
Design. To not have an overly complex design, the number of patients N was fixed at 60. Additionally, the true number of source signals per cluster \(Q^\text {true}\) was set at 20, the number of voxels V at 1000 and finally the number of time points T at 100. Note that these settings are commonly encountered in an fMRI study, except than for the number of voxels, which is often (much) larger.Footnote 4 Furthermore, the following data characteristics were systematically varied in a completely randomized four-factorial design, with the factors considered as random:
-
The true number of clusters, K, at two levels: 2 and 4;
-
The cluster sizes, at two levels: equally sized and unequally sized clusters;
-
The degree of overlap among clusters, at 5 levels: small, medium, large, very large and extreme;
-
The percentage of noise in the data, at four levels: 10%, 30%, 60% and 80%.
Data generation procedure. The data were generated as follows: first, a common set of independent source signals \({\mathbf {S}}^\text {base}\) (\(1000 \times 20\)) was generated where each source signal was simulated from \(U(-1,1)\). Next, depending on the desired number of clusters K, 2 or 4 temporary matrices \({\mathbf {S}}^\text {temp}_{k}\) (\(k=1,\dots ,K\)) were generated from \(U(-1,1)\). To obtain the cluster specific source signals \({\mathbf {S}}^k\), weighted \({\mathbf {S}}^\text {temp}_{k}\)’s were added to \({\mathbf {S}}^\text {base}\): \({\mathbf {S}}_k = {\mathbf {S}}^\text {base} + w {\mathbf {S}}^\text {temp}_{k}\) (for \(k=1,\dots ,K\)). Note that by varying the value of w, the cluster overlap factor was manipulated. In particular, a pilot study indicated that a weight of 0.395, 0.230, 0.150, 0.120 and 0.080 results -for \(K=4\)- in an average pairwise modified RV-coefficient between the cluster specific \({\mathbf {S}}_k\)’s of , respectively, 0.75 (small overlap), 0.90 (medium overlap), 0.95 (large overlap), 0.97 (very large overlap) and 0.99 (extreme overlap).
Next, for each patient i (\(i = 1,\dots ,60\)), patient specific time courses \({\mathbf {A}}_i\) (\(100 \times 20\)) were generated by simulating fMRI time courses using the R package neuRosim (Welvaert et al. 2011). Here, the default settings of neuRosim were used, that is, the repetition time was set at 2.0 s and the baseline value of the time courses equalled 0. Note that the neuRosim package ensures that the fluctuations of the frequencies in the time courses are between 0.01 and 0.10 Hz, which is a frequency band that is relevant for fMRI data (Fox and Raichle 2007).
To obtain noiseless fMRI data \({\mathbf {Z}}_i\) for each patient i (\(i = 1,\dots ,60\)), a true clustering of the patients was generated and the patient specific time courses \({\mathbf {A}}_i\) were linearly mixed by one of the cluster-specific source signals \({\mathbf {S}}_k\). More specifically, if equally sized clusters were sought for, the 60 patients were divided into clusters containing exactly \(\frac{60}{K}\) patients; for conditions with unequally sized clusters, the patients were split up into two clusters of 15 and 45 patients (\(K = 2\)-conditions) or into four clusters of size 5, 10, 20 and 25 patients (\(K = 4\)-conditions). For each patient, its \({\mathbf {A}}_i\) was multiplied with the \({\mathbf {S}}_k\) corresponding to the cluster to which the patient in question was assigned to.
Finally, noise was added to each patient’s true data \({\mathbf {Z}}_i\). To this end, first, a noise matrix \({\mathbf {E}}_i\) (\(1000 \times 100\)) was generated for each patient i by independently drawing entries from \({\mathcal {N}}(0,1)\). Next, the matrices \({\mathbf {E}}_i\) were rescaled such that their sum of squared entries (SSQ) equalled the SSQ of the corresponding \({\mathbf {Z}}_i\). Finally, a weighted version of the rescaled \({\mathbf {E}}_i\) was added to \({\mathbf {Z}}_i\) to get data with noise \({\mathbf {X}}_i\): \({\mathbf {X}}_i = {\mathbf {Z}}_i + w{\mathbf {E}}_i = {\mathbf {S}}_k({\mathbf {A}}_i)^T + w{\mathbf {E}}_i\). The weight w was used to manipulate the amount -percentage- of noise in the data. In particular, the desired percentage of noise can be obtained by taking \(w = \sqrt{{\frac{\mathrm{noise}}{1-{\mathrm{noise}}}}}\), with \(\text {noise}\) equalling 0.10, 0.30, 0.60 and 0.80 for the 10%, 30%, 60% and 80%-noise condition, respectively.
Data analysis. For each cell of the factorial design, 10 replication data sets were generated. Thus, in total, 2 (number of clusters) \(\times \) 2 (cluster sizes) \(\times \) 5 (cluster overlap) \(\times \) 4 (noise level) \(\times \) 10 (replications) = 800 data sets \({\mathbf {X}}_{i}\) (\(i=1, \dots , 60\)) were simulated. Each \({\mathbf {X}}_{i}\) was subjected to (1) a single-subject ICA and (2) a single-subject PCA, both dimension reduction methods with \(Q^\text {true}= 20\) components, yielding estimated source signals \(\hat{{\mathbf {S}}}_{i}^\text {ICA}\)/\(\hat{{\mathbf {S}}}_{i}^\text {PCA}\) (see bottom panel of Fig. 3). Notice that only ICA/PCA with the true number of components \(Q^\text {true}\) (i.e., the number of components used to generate the data) was performed as model selection is a non-trivial and complex task that falls outside the scope of this paper which focuses on clustering patients. Next, for each data set, a (dis)similarity matrix was computed (see Sect. 3.2) using both the original data \({\mathbf {X}}_{i}\) as well as the ICA/PCA reduced data \(\hat{{\mathbf {S}}}_{i}^\text {ICA}\)/\(\hat{{\mathbf {S}}}_{i}^\text {PCA}\). Finally, each (dis)similarity matrix was analysed with each of the following clustering methods using only the true number of clusters K: (1) AP, (2) PAM, (3) HC using Ward’s method and (4) HC using complete linkage. Note that we only analysed the data with the true number of clusters K as selecting the optimal number of clusters is a difficult task that exceeds the goals of this study. Moreover, several approaches have been proposed and evaluated in the literature to tackle this vexing issue (Milligan and Cooper 1985; Rousseeuw 1987; Tibshirani et al. 2001).
As Clusterwise SCA-ECP (De Roover et al. 2012) simultaneously performs dimension reduction and clustering, it can be considered as a competitor of the proposed two-step procedure.Footnote 5 In order to investigate the cluster recovery performance of Clusterwise SCA-ECP, we selected a relatively easy and a fairly difficult condition from the simulation design. More specifically, we took the 10 data sets from the simulation condition with 2 equally sized clusters, with a medium overlap (RV = .90) and either 60% noise or 80% noise added to the data. We analysed these data sets with Clusterwise SCA-ECP using publicly available software (De Roover et al. 2012) and choosing \(K=2\) and \(Q=20\). As the Clusterwise SCA-ECP loss function suffers from local minima, a multi-start procedure with 25 random starts was implemented.
Over- and underestimation of the true number of ICA components\(Q^\text {true}\). To study the effect of under- and overestimating the true number of ICA components \(Q^\text {true}\) on the cluster recovery performance, we selected the same relatively easy and fairly difficult condition from the simulation design as used for the Clusterwise SCA-ECP analysis (see above). Next, we applied the proposed two-step procedure (using ICA based data reduction and all clustering methods) to the data sets from the selected simulation conditions using a range for Q (which was kept equal across patients) such that Q was lower (underestimation) and larger (overestimation) than the true number of components \(Q^\text {true}\). In particular, we analysed these data sets using Q=2–40 (in steps of 2) components (remember that \(Q^\text {true}\) equals 20) and \(K=2\) clusters, which equals the true number of clusters used to generate the data.
Software. All simulations were carried out in R version 3.4 (Core Team 2017) on a high-performance computer cluster that enables computations in parallel. In order to compute the modified RV-coefficients, the R package MatrixCorrelation was used (Indahl et al. 2016). For ICA, the function icafast from the R package ica was adopted (Helwig 2015). The AP method was carried out with the apclusterK function from the R package apcluster (Bodenhofer et al. 2011). Note that this function has the option to specify a desired number of clusters K, which can be obtained by playing around with the ‘preference’ values for the objects (see Sect. 3.3). PAM was performed using the function pam from the cluster package (Maechler et al. 2017). Finally, both hierarchical clustering methods were executed with the hclust function from the Rstats package.
4.3 Results
To evaluate the recovery of the true cluster structure, the Adjusted Rand Index (ARI; Hubert and Arabie 1985) between the true patient partition and the patient partition estimated by the two-step procedure was computed. The ARI equals 1 when there is a perfect cluster recovery and 0 when both partitions agree at chance level.
The overall results -averaged across all generated data sets- show that the hierarchical clustering methods (ARI Ward = 0.63, ARI complete linkage = 0.54) outperform both AP (ARI = 0.48) and PAM (ARI = 0.48). Table 1 displays the mean ARI value for each level of each manipulated factor per clustering method and this for when the original data, the PCA reduced data and the ICA reduced data are used as input for the clustering. From this table, it appears that for each level of each factor, hierarchical clustering using Ward’s method yields the largest ARI value among the clustering methods, and this for each reduction method (none/PCA/ICA) used. Further, for each clustering method, a high recovery of the cluster structure occurred when the cluster overlap was small. However, albeit to a lesser extent for the hierarchical clustering method with Ward’s method, these results deteriorate when the overlap between cluster structures increases. Additionally, recovery performance becomes worse when the data contain a larger number of clusters that are of unequal size and when the data are noisier.
Comparing clustering original data and PCA reduced data to clustering ICA reduced data, a large recovery difference in favour of ICA reduced data is observed. In particular, all clustering methods recover the cluster structure to a relatively large extent when using ICA reduced data (mean overall ARI \(> 0.70\)), whereas recovery is much worse when no ICA reduction takes place (mean overall ARI’s between 0.37 and 0.55). This implies that when the cluster structure is determined by similarities and differences in independent non-Gaussian components underlying the observed data, these clusters cannot be recovered well by clustering the observed data. Moreover, reducing the data with PCA before clustering does not lead to an improved cluster recovery. Indeed, ICA needs to be applied first to reveal the true clusters (for a further discussion of this implication, see the Discussion section). Just as was the case for non-reduced data and PCA reduced data, for ICA reduced data -although to a smaller extent- recovery decreases when the data contain more noise and more clusters exist in the data that show larger overlap.
To evaluate potential main and interaction effects between the manipulated factors, a mixed-design analysis of variance (ANOVA) was performed using Greenhouse-Geisser correction to account for violations of the sphericity assumption (Greenhouse and Geisser 1959). Here, the ARI was used as the dependent variable, the aforementioned factors pertaining to data characteristics (see Sect. 4.2) were treated as between-subjects factors and the clustering method (four levels) and the type of data reduction (i.e., ICA, PCA or none) applied before clustering (three levels) were used as within-subjects factors. When discussing the results of this analysis, only significant effects with a medium effect size as measured by the generalized eta squared \(\eta _\text {G}^2\) (Olejnik and Algina 2003) are considered (i.e., \(\eta _\text {G}^2 > 0.15\)). The generalized eta squared was adopted as effect size because for complex designs such as the current one this effect size is more appropriate to use than the ordinary—partial—eta squared effect size measure (Bakeman 2005).
The ANOVA table resulting from the analysis is presented in Table 2, only displaying main and interaction effects with an effect size \(\eta _\text {G}^2 > 0.15\) (and \(p< 0.05\)). From this table, it appears that cluster recovery mainly depends on the amount of cluster overlap (\(\eta _\text {G}^2 = 0.91\)), the amount of noise present in the data (\(\eta _\text {G}^2 = 0.78\)), the type of data reduction (\(\eta _\text {G}^2 = 0.70\)) and the clustering method used (\(\eta _\text {G}^2 = 0.29\)). In particular, as can be seen also in Table 1, cluster recovery deteriorates when clusters overlap more (mean ARI of 0.96, 0.77, 0.44, 0.30 and 0.19 for small, medium, large, very large and extreme cluster overlap, respectively), when the noise is the data increases (mean ARI of 0.67, 0.67, 0.56 and 0.24 for 10%, 30%, 60% and 80% of noise, respectively), when no ICA reduction is performed before clustering (mean ARI of 0.74, 0.43 and 0.43 for ICA reduced, PCA reduced and non-reduced data, respectively) and when AP and PAM are used compared to when HC is used (mean ARI of 0.63, 0.54, 0.48 and 0.48 for Ward’s HC, complete linkage HC, PAM and AP, respectively). These main effects, however, are qualified by three two-way interactions. In particular, the amount of cluster overlap interacts both with the type of data reduction (\(\eta _\text {G}^2 = 0.57\)) and the amount of noise in the data (\(\eta _\text {G}^2 = 0.49\)): the decrease in cluster recovery with increasing cluster overlap is more pronounced when the data are not reduced with ICA—compared to when data are ICA reduced—and when there is more noise present in the data. The third two-way interaction effect refers to the interaction between the amount of noise in the data and the type of data reduction (\(\eta _\text {G}^2 = 0.53\)). In particular, the cluster recovery is less affected by increasing amounts of noise after ICA reduction compared to when no ICA reduction is applied. Finally, two sizeable three-way interactions qualify the above discussed effects. The first three-way interaction involves the amount of cluster overlap, the amount of noise in the data and the type of data reduction (\(\eta _\text {G}^2 = 0.46\)). In Fig. 4, which displays this three-way interaction effect, one can clearly see that the detrimental combined effect of larger cluster overlap and more noisy data is way more pronounced when the data are not reduced (upper panel of Fig. 4) or PCA reduced (middle panel) before clustering compared to when ICA reduction was applied (lower panel) before clustering.
Mean Adjusted Rand Index (ARI) as a function of the degree of cluster overlap (X-axis), the amount of noise (colours) and the data reduction method (top: no reduction; middle: after PCA reduction; bottom: after ICA reduction). ICA independent component analysis, PCA principal component analysis (colour figure online)
The second three-way interaction refers to the amount of overlap between clusters, the amount of noise in the data and which particular clustering method is used (\(\eta _\text {G}^2 = 0.19\)). This three-way interaction is presented in Fig. 5 in which it clearly can be seen that the cluster recovery deteriorates when both the extent of cluster overlap and the amount of noise increases, with this effect being less pronounced for the hierarchical clustering using Ward’s method compared to the other clustering methods.
Mean Adjusted Rand Index (ARI) as a function of the degree of cluster overlap (X-axis), the amount of noise (colours) and the clustering method (four panels). AP affinitiy propagation, HCC hierarchical clustering with complete linkage, HCW hierarchical clustering with Ward’s method, PAM partitioning around medoids (colour figure online)
Clusterwise SCA-ECP. Table 3 displays the mean ARI value for each cluster method after either no reduction, PCA data reduction or ICA data reduction for a fairly easy condition and a fairly difficult condition of the simulation design. Additionally, also the mean ARI values for the results obtained with a Clusterwise SCA-ECP applied to the data sets from the same two conditions are shown. For both conditions, the mean ARI obtained with Clusterwise SCA-ECP (mean ARI of 0.59 and 0.11 for the easy and difficult condition, respectively) is substantially lower than the ARI’s resulting from any of the clustering methods in combination with ICA or PCA reduction before clustering. Even after no reduction at all, the ARI’s of the clustering methods outperform Clusterwise SCA-ECP. All in all, the results of these two conditions suggest that Clusterwise SCA-ECP fails in recovering the clustering structure underlying the data.
Over- and underestimation of\(Q^\text {true}\). Figure 6 displays how the recovery performance (in terms of ARI) is affected by choosing different values of Q for the ICA reduction (note that for each patient \(Q^\text {true}=20\)) for each of the four clustering methods and this when looking at a relatively easy (left panel of Fig. 6) and a fairly difficult (right panel) condition of the simulation design. The results clearly indicate that for the relatively easy condition with 60% noise all methods perform excellent when Q (kept equal across patients) is equal to or larger than \(Q^\text {true}\), which implies that overestimation of Q in this condition does not have a detrimental effect on cluster recovery. For the difficult condition with 80% noise, Ward’s hierarchical clustering method clearly outperforms the other clustering methods when Q is equal to or larger than \(Q^\text {true}\). Overestimation of Q has a small positive effect on cluster recovery for PAM and AP, but a less univocal effect for the hierarchical clustering methods. When Q is underestimated, cluster recovery declines, with this decline being stronger the more Q is lower than \(Q^\text {true}\). In the relatively easy condition especially AP and PAM are affected by under-estimation (i.e., the decline in recovery starts already at \(Q= 16\)), whereas hierarchical clustering with complete linkage and to a stronger extent Ward’s hierarchical clustering are almost not affected by small to moderate amounts of underestimation (i.e., both methods perform well as long Q is equal or larger than 10). This implies that in this easy condition both hierarchical clustering methods need fewer ICA components to arrive at a good clustering solution than AP and PAM. In the difficult condition, however, all clustering methods suffer from underestimation of Q, and this even for small amounts of underestimation. Note that Ward’s hierarchical clustering in the difficult condition outperforms all other clustering methods, except when \(Q=2\).
Mean Adjusted Rand Index (ARI) as a function of the number of retained ICA components Q (X-axis) and clustering method (colours) for two design conditions (two equally sized clusters with medium cluster overlap and 60%—left panel—or 80%—right panel—of noise in the data). AP affinitiy propagation, HCC hierarchical clustering with complete linkage, HCW hierarchical clustering with Ward’s method, PAM partitioning around medoids (colour figure online)
5 Illustrative application
5.1 Motivation and data
In this section, the proposed two-step procedure will be illustrated on an empirical multi-subject resting-state (i.e., patients were scanned while not engaging in a particular task) fMRI data set concerning dementia patients. Although these patients are diagnosed with either Alzheimer’s disease (AD) or behavioral variant frontotemporal dementia (bvFTD), these existing labels will not be taken into account when performing the cluster analysis. Previous studies that investigated the brain dysfunctions in these patient groups took the (labels for the) subtypes for granted, often focussed on a priori defined brain regions and mainly tried to discriminate each patient subtype from healthy controls. For example, it has been shown that -compared to healthy subjects- decreased activity occurs in the default mode network (i.e., medial prefrontal cortex, posterior cingulate cortex and the precuneus) in AD patients (Greicius et al. 2004) and in the salience network (i.e., the anterior insula cortex and dorsal anterior cingulate cortex) for bvFTD patients (Agosta et al. 2013). However, by selecting a priori defined regions, analyses may overlook potentially relevant brain areas or FC patterns that are involved in these pathologies. Moreover, by focusing on the given dementia subtypes, the heterogeneity within each subtype is not accounted for. This within-subtype heterogeneity may point at the need for a further subdivision of the subtypes. As a solution, a data-driven whole-brain clustering method may be adopted that allocates patients to homogenous groups based on similarities and differences in whole-brain (dys)functioning. As such, heterogeneity in brain functioning within patient clusters is decreased. This may in an explorative way guide hypotheses about unknown subtypes of dementia, which should be later tested in confirmatory studies. Moreover, relevant but yet uncovered FC patterns that are able to differentiate between -known and unknown- dementia subtypes may get disclosed.
The goal of the current application in which the proposed two-step procedure is applied to multi-subject fMRI data regarding dementia patients is to cluster the patients on the basis of similarities and differences in the FC patterns underlying their data. It has to be stressed that this analysis is performed without using any information about the diagnostic labels (i.e., the analysis is completely unsupervised). After clustering the patients, however, the diagnostic labels will be used to interpret and validate the derived clustering. To further validate the obtained clustering, an ad hoc procedure is adopted in which the FC patterns in each cluster are matched to template FC patterns, allowing to identify the dementia related FC patterns within each cluster (and differences therein between clusters). Such an ad hoc procedure is needed as fMRI data are very noise and ICA often retains FC patterns that represent (systematic) noise aspects of the data that are not physiologically or biologically relevant for dementia (e.g., head motion artefacts). Note that validating extracted ICA components is still a vexing issue within the fMRI community. We acknowledge that a more rigorous validation procedure is needed to demonstrate the existence of a novel –yet undiscovered– subtype of dementia. We believe, however, that such a rigorous validation goes beyond the scope of the current study which mainly wants to present a method to cluster patients based on FC patterns.
The data set includes fMRI data for 20 patients, of which 11 are diagnosed as AD patients and 9 as bvFTD patients. The data for each patient contains 902,629 voxels measured at 200 time points (i.e., a 6 minute scan). The data acquisition, pre-processing (e.g., accounting for movement during a scanning session) and registration procedure (i.e., the registration of each patient’s brain to a common coordinate system of the human brain, see Mazziotta et al. 2001) are fully described in Hafkemeijer et al. (2015).Footnote 6
5.2 Data analysis
To make the analysis more feasible, we downsampled the voxels of the data \({\mathbf {X}}_{i}\) of each patient such that the spatial resolution of each voxel is reduced from 2 \(\times \) 2 \(\times \) 2mm to 4 \(\times \) 4 \(\times \) 4mm. This procedure is performed using the subsamp2 command from the FMRIB Software Library (FSL, version 5.0; Jenkinson et al. 2012). As such, the number of voxels in each data set \({\mathbf {X}}_{i}\) is reduced from 902,629 to 116,380. Additionally, a brain mask was applied to each data set such that only the voxels that are present within the brain are included in the analysis, resulting in a further reduction of the number of voxels to 26,647 voxels. As a result, the data used for the analysis contains 20 (patients) \(\times \) 26,647 (voxels) \(\times \) 200 (time points) = 106,588,000 data entries.
To analyze the data, as a first step, the dimensionality of the data \({\mathbf {X}}_{i}\) of each patient was reduced by means of ICA. To this end, the icafast function of the R-package ica (Helwig 2015) was used, and 20 ICA components were retained for each patient. We opted for this number of components since this is an often used model order for resting-state fMRI data that usually results in FC patterns that show a close correspondence to the brain’s known architecture (Smith et al. 2009). Next, a dissimilarity matrix was computed by calculating for each patient pair one minus the modified RV-coefficient between the ICA components \(\hat{{\mathbf {S}}}_i\) of the members of the patient pair (see the procedure described in Sect. 3.2). In a second step, a hierarchical cluster analysis using Ward’s method was performed on this dissimilarity matrix and the resulting dendrogram was cut at a height such that \(K= 2, 3\) and 4. Due to the small number of patients (i.e., 20) in the data set, it did not make much sense to check for larger numbers of clusters.
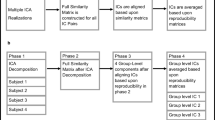
To validate the obtained clustering, the associated cluster specific FC patterns were investigated. To this end, for each cluster separately, ICA was performed on the temporally concatenated data sets of the patients belonging to that cluster (i.e., the original data \({\mathbf {X}}_{i}\) of all patients belonging to a particular cluster were concatenated in the temporal dimension). The latter method is known in the literature as group ICA (Calhoun et al. 2009) and yields common FC patterns that are representative for a group of patients. As the ICA components have no natural order, identifying cluster-specific ICA components related to dementia is a challenging task, which is even further complicated by the large amount of noise that is present in fMRI data and the fact that ICA may retain irrelevant noise components (e.g., motion artifacts). To identify dementia related FC patterns and investigate how these patterns differ between clusters, the FC patterns of the obtained clusters were matched on a one-to-one basis. To this end, the obtained FC patterns in each cluster were compared to a reference template consisting of eight known FC patterns, which encompass important visual cortical areas and areas from the sensory-motor cortex (see left part of Fig. 8). These template FC patterns have been encountered consistently in many -mainly healthy- subjects (Beckmann et al. 2005), but disruptions in these FC patterns have also been shown to be related to several mental disorders (i.e., depression and other psychiatric disorders, see Greicius et al. 2004; Gour et al. 2014). For each template FC pattern in turn, the ICA FC pattern in each cluster was determined that has the largest absolute Tucker congruence coefficient (Tucker 1951) with the template FC pattern in question. Note that whereas the modified RV coefficient is often adopted to assess the similarity of matrices (see Sect. 3.2), the Tucker congruence coefficient is a more natural measure to quantify the similarity of vectors (with a Tucker congruence value larger than .85 being considered as indicating a satisfactory similarity between vectors, see Lorenzo-Seva and Ten Berge 2006). As such, FC patterns from different clusters were matched to each other in a one-to-one fashion (for an example, see Fig. 8).
To evaluate the proposed validation strategy based on group ICA and template FC patterns, also a group PCA-based validation strategy was performed. In particular, for each cluster, the data sets of the patients belonging to that cluster were temporally concatenated and analyzed with PCA. The same matching strategy adopting template FC patterns was used to match PCA components across clusters in a one-to-one fashion. As PCA, as opposed to ICA, has rotational freedom, the matched PCA components in each cluster were optimally transformed towards the template FC patterns by means of a Procrustes (oblique) rotation.
Further, we investigated the stability of the obtained cluster solution by employing a bootstrap procedure described in Hennig (2007) and implemented in the R package fpc (Hennig 2018). This procedure uses the Jaccard coefficient, which quantifies the similarity between sample sets (e.g., two clusterings), as a measure of cluster stability. The cluster stability is assessed by computing the average maximum Jaccard coefficient over bootstrapped dataset, with the maximum being taken to account for permutational freedom of the clusters. Here, each cluster receives a bootstrapped mean Jaccard value which indicates whether a cluster is stable (i.e., a value of .85 or above) or not.
Finally, for comparative purposes, we (1) analysed the data with Clusterwise SCA-ECP (with K\(=\) 2 clusters and Q\(=\) 20 components), (2) applied ICA (with Q\(=\) 20) in combination with the other clustering methods (with K\(=\) 2) and (3) performed an analysis with PCA (instead of ICA) reduction with Q\(=\) 20 and an analysis without ICA reduction (i.e., clustering the \({\mathbf {X}}_{i}'s\) directly) before clustering (with always K\(=\) 2 clusters). To perform Clusterwise SCA-ECP, we used the Matlab code of De Roover et al. (2012) and employed a multi-start procedure with 25 starts to avoid suboptimal cluster solutions representing local minima of the Clusterwise SCA-ECP loss function.
5.3 Results
Cluster solution. Table 4 shows the clustering obtained with the proposed two-step procedure using ICA with Ward’s HC for \(K = 2\), 3 and 4 clusters. The two-cluster solution yields two more or less equally sized clusters, whereas the three- and four-cluster solution results in some clusters being very small. In particular, compared to the two-cluster solution, the three-cluster solution places three patients in a separate cluster (i.e., AD8, AD9 and AD11), whereas the four-cluster solution on top places a single patient into a separate cluster (i.e., FTD8). Since the cluster solutions with \(K = 3\) and \(K = 4\) have clusters with very few members (e.g., singleton cluster 4 for \(K = 4\)) and investigating the FC patterns underlying such small clusters might not be informative at all, only the solution with two clusters seems to make sense and will therefore be further discussed.
The two-cluster solution of the two-step procedure using ICA reduction and Ward’s method is displayed in Fig. 7 as a dendrogram, which is cut at a height of 1 resulting in a partition of the patients into K\(=\) 2 clusters (indicated by lightgrey dashed rectangles). As can be seen in this figure (and also from the second column of Table 4/5), eight patients were allocated to the blue cluster (indicated by the blue branches), whereas the red cluster (indicated by the red branches) contains 12 patients. Regarding the stability of the obtained two-cluster solution, the blue and red cluster yielded a bootstrapped mean Jaccard coefficient of 0.87 and 0.85, respectively. This indicates that both clusters are stable.
Dendrogram obtained after applying the two-step procedure with ICA with \(Q = 20\) and Ward’s hierarchical clustering to the illustrative multi-subject fMRI data set regarding dementia patients. The tree is cut such that K\(=\) 2 clusters are retained, which are indicated with branches in a different colour (blue vs. red) and by lightgrey dashed rectangles. The obtained clustering is compared with the existing patient labels: bvFTD patients are indicated with golden nodes, whereas AD patients are indicated with green nodes. The distance at which patients/patient clusters are merged is indicated on the Y-axis (colour figure online)
Table 5 shows the partitions with \(K = 2\) obtained for each type of data reduction (either using ICA reduction, PCA reduction or no reduction) in combination with each of the four clustering methods and for Clusterwise SCA-ECP. The obtained partitions with the four clustering methods after both ICA reduction and no reduction show similar results. In particular, when comparing two partitions at most five patients (but often only one or two) switch their cluster, with the patients that switch always belonging to the same cluster in both partitions. However, for the PCA reduction method, only the partition obtained with Ward’s HC equals one of the above mentioned partitions (i.e., the partition obtained with AP and complete linkage hierarchical clustering after no reduction). The partitions from PAM, AP and the hierarchical clustering using complete linkage, all do not make much sense as they allocate 17 patients to one cluster and only 3 patients to the second cluster. Finally, the partition obtained with Clusterwise SCA-ECP does not seem to be related to any of the obtained partitions.
To validate the obtained two-cluster solution, it was compared to the diagnostic labels by means of the ARI coefficient (see Sect. 4.3) and the balanced accuracy, which is often used to indicate classification performance (García et al. 2009). It appears, as can be seen in the dendrogram in Fig. 7, that the blue cluster mainly consists of bvFTD patients (i.e., six out of eight) which are indicated with golden nodes, whereas nine out of twelve of the patients in the red cluster are diagnosed with AD (green nodes). It can be concluded that the obtained clusters only partially correspond with the diagnostic labels. In particular, the balanced accuracy of the obtained clustering is 0.75 and the ARI equals 0.21. This demonstrates that differences in FC patterns do not fully correspond with the known dementia subtypes. This is in line with previous research that showed that supervised learning methods can predict dementia subtype membership to an acceptable large extent—but not perfectly—based on fMRI information alone (de Vos et al. 2018). Clusterwise SCA-ECP yields a clustering that does not capture the actual diagnosis of the patients at all: balanced accuracy \(=\) 0.51 and ARI \(=\) −0.05, which for both measures are at chance level (also see Table 5). Compared to ICA with Ward’s HC, ICA with single linkage hierarchical clustering recovers the diagnostic labels to the same extent (balanced accuracy \(=\) 0.77 and ARI \(=\) 0.21), whereas the diagnostic labels are disclosed to a bit lesser extent by ICA with AP and PAM (balanced accuracy \(=\) 0.70 and ARI \(=\) 0.12). When applying the clustering methods after a PCA reduction, only Ward’s HC yields a clustering that corresponds to the diagnostic labels on an above chance level (balanced accuracy \(=\) 0.75 and ARI \(=\) 0.21), whereas the other three clustering methods after PCA reduction do not capture the diagnostic labels at all (all ARI’s \(=\) − 0.02 and balanced accuracy’s \(=\) 0.25). Remarkably, when applying the clustering methods without a dimension reduction, the results are very similar to the results with ICA reduction. In particular, the balanced accuracy and ARI values are 0.75 and 0.21 for all four clustering methods. Note, however, that an additional analysis using an ICA reduction with Q=5 components (instead of 20) resulted in a balanced accuracy of 0.80 and an ARI of 0.33 for Ward’s hierarchical clustering (see Table 5). This suggests that taking a smaller number of components than the initial chosen number of \(Q = 20\) components, may result in a partition that better captures the two existing patient groups.
Cluster specific FC patterns. To interpret and compare the underlying FC patterns for the two clusters to each other, in Fig. 8, we plotted each template FC pattern (left part of the figure) against the component in the first (middle part, in red) and second (right part, in blue) cluster (obtained with the two-step approach with ICA and Ward’s hierarchical clustering) which resembles the template FC pattern in question the most. As such, a one-to-one mapping of the FC patterns in both clusters is obtained. As can be seen in Fig. 8, qualitative differences between (some) FC patterns of both clusters exist. For example, a clear difference is visible between the estimated salience network (template C in Fig. 8) of the blue cluster (middle panel, mainly bvFTD) and red cluster (right panel, mainly AD). In particular, the salience network seems more devastated for bvFTD (blue cluster) than for AD (red cluster) patients. This result is in line with previous research that demonstrated a diminished activity within the salience network especially for bvFTD patients (Agosta et al. 2013). Further, for some templates, like the right and left frontotemporal network (template G and H), a rather similar FC pattern is found for AD patients but not for bvFTD patients, implying that these FC patterns are less pronounced for bvFTD patients than for AD patients. For the default mode network (template E), however, no clear differences between both clusters are visible, which contrasts with previous findings.
Cluster specific FC patterns (matched to a template in terms of Tucker congruence value) for the clusters obtained with the two-step procedure (ICA with Ward’s hierarchical clustering) applied to the dementia data. The characteristic FC patterns for each cluster are estimated by performing Group ICA on the data per cluster. In the left panel (yellow), eight known FC patterns from a template from Beckmann et al. (2005) are displayed (in the rows). Note that each FC pattern is presented with three pictures as the brain can be displayed in three different planes: the sagittal plane (first picture), the coronal plane (second picture) and the axial plane (third picture). In the middle panel (blue) the eight estimated FC patterns, which are most similar to the template patterns, for the blue cluster (mainly BvFTD patients) are displayed. In the right panel (red), the eight estimated FC patterns -matched to the templates- for the red cluster (mainly AD patients) are visualised. The eight known ’template’ FC patterns refer to eight important brain networks: a medial visual network, b lateral visual network, c salience network, d sensorimotor network, e default mode network, f executive control network, g right frontotemporal network and h left frontotemporal network (colour figure online)
Table 6 displays, for each template FC pattern, the absolute Tucker congruence coefficient between the template in question and the matched FC pattern from each cluster. From this table, one can see that, overall, the two-step procedure with Ward’s hierarchical clustering finds FC patterns that match the template FC patterns to some extent (a mean Tucker congruence of 0.49 and 0.52 for the blue and red cluster, respectively).
The cluster specific FC patterns obtained with Clusterwise SCA-ECP before (left panels) and after an orthogonal rotation with varimax (right panels) are displayed in Fig. 9. In this figure, it clearly can be seen that Clusterwise SCA-ECP does not yield FC patterns that are related to dementia or to relevant differences between dementia subtypes. In particular, Clusterwise SCA-ECP yields FC patterns that encompass too large brain regions, which can be a consequence of Clusterwise SCA-ECP only enforcing orthogonality but not spatial independence and non-Gaussianity on the FC patterns. Moreover, as can be seen in Table 6, the cluster specific FC patterns obtained with (an unrotated) Clusterwise SCA-ECP match the template FC patterns to a smaller extent (a mean Tucker congruence of 0.35 and 0.41 for cluster 1 and 2, respectively) than the FC patterns from the two-step procedure (i.e., ICA with Ward’s HC followed by group ICA). This result is also true after a varimax rotation of the Clusterwise SCA-ECP solution (i.e., mean Tucker congruence of 0.37 and 0.42 for cluster 1 and 2, respectively).
Cluster specific FC patterns (matched to a template) obtained from the Clusterwise SCA-ECP analysis applied to the dementia data. Note that each FC pattern is presented with three pictures as the brain can be displayed in three different planes: the sagittal plane (first picture), the coronal plane (second picture) and the axial plane (third picture). The first two columns (one for each cluster) refer to the unrotated FC patterns obtained with the Clusterwise SCA-ECP analysis. The last two columns show varimax rotated FC patterns of the Clusterwise SCA-ECP solution. Each FC pattern (in the rows) is optimally matched in terms of Tucker congruence value to one of the eight known FC patterns (displayed in Fig. 8) from a template from Beckmann et al. (2005). The eight known ’template’ FC patterns refer to eight important brain networks: a medial visual network, b lateral visual network, c salience network, d sensorimotornetwork, e default mode network, f executive control network, g right frontotemporal network and h left frontotemporal network (colour figure online)
Applying a Group PCA (instead of Group ICA) based validation strategy to the clusters obtained with the two-step procedure (ICA with Ward’s HC) resulted in FC patterns (not shown) that have no clear physiological meaning. Moreover, the obtained FC patterns match the template patterns to a smaller extent than the patterns resulting from Group ICA (i.e., a mean Tucker congruence of 0.34 for both clusters). Optimally (obliquely) rotating these FC patterns towards the template patterns did not substantially improve the FC patterns overall (in Table 6: mean Tucker values of 0.35 and 0.37) and also did not result in meaningful FC patterns (see right panels of Fig. 10). Fully exploiting the rotational freedom of Clusterwise SCA-ECP (left panels of Fig. 10) by transforming the SCA-ECP components in an optimal way towards the template FC patterns by means of a Procrustes (oblique) rotation does not lead to cluster specific FC patterns that are easy to interpret or that are physiologically meaningful. The mean Tucker congruence values, however, become a little bit larger than those associated with the FC patterns from Group ICA (i.e., 0.52 and 0.56). This indicates that a large(r) Tucker congruence value alone is not enough to consider a FC pattern as substantively meaningful. Looking at Tucker congruence values, however, can help to identify the FC patterns that are worth for further investigation.
Cluster specific FC patterns (matched to a template) of the Clusterwise SCA-ECP analysis after Procrustes rotation and the two-step procedure (ICA with Ward’s hierarchical clustering) with Group PCA and Procrustes rotation applied to the dementia data. Note that each FC pattern is presented with three pictures as the brain can be displayed in three different planes: the sagittal plane (first picture), the coronal plane (second picture) and the axial plane (third picture). The second and third columns (one for each cluster) refer to the Procrustes rotated FC patterns obtained with the Clusterwise SCA-ECP analysis. The last two columns show the FC patterns obtained by applying Group PCA with Procrustes rotation to the clusters from the two-step procedure (ICA with Ward’s HC). Each FC pattern (in the rows) is optimally matched in terms of Tucker congruence value to one of two known FC patterns from a template from Beckmann et al. (2005). After this matching procedure, the FC patterns are optimally rotated to the template FC patterns using a Procrustes (oblique) rotation. The two known ’template’ FC patterns refer to two important brain networks relevant for dementia: (1) default mode network (template E in Fig. 8; top row) and (2) salience network (template C in Fig. 8; bottom row)
6 Discussion
A new avenue in neuroscientific research pertains to clustering patients based on multi-subject fMRI data to, for example, obtain a categorization of mental disorders (e.g., dementia and depression) in terms of brain dysfunctions. This search for a brain-based categorization of mental disorders, which fits nicely within the promising trend of personalized/precision psychiatry (Fernandes et al. 2017), may result in improved treatments and outcomes for patients and enhance the early detection of patients at risk (Insel and Cuthbert 2015). However, due to the three-way nature of multi-subject fMRI data, most commonly adopted clustering methods cannot be used to this end in a straightforward way. Therefore, in this article, a two-step procedure was proposed that consists of (1) reducing the data with ICA and (2) clustering the patients into homogenous groups based on the (dis)similarity between patient pairs in terms of ICA components/FC patterns as measured by the modified RV-coefficient.
An extensive simulation study showed that our two-step procedure adopting ICA and Ward’s hierarchical clustering performs well in general and is superior to the two-step procedure using ICA with one of the other clustering methods (i.e., AP, PAM and complete linkage hierarchical clustering). The cluster recovery obtained with Ward’s hierarchical clustering is excellent to good, except when clusters overlap to a very large extent and the data contains large amounts of noise. Further, not reducing the data with ICA or reducing the data with PCA prior to clustering seriously affects the cluster recovery and this especially for AP and PAM. When the overlap between clusters is large and/or when the data are very noisy, the performance of AP, PAM and -to a lesser extent- complete linkage hierarchical clustering deteriorates seriously, whereas hierarchical clustering with Ward’s method is less affected. Moreover, these effects are way more pronounced when the original data are clustered without performing first an ICA reduction or when the data are reduced with PCA before clustering. Interestingly, when the clusters have a small overlap, Ward’s hierarchical clustering also performs excellent when PCA reduction or no ICA reduction is performed before clustering, with this to a smaller extent being true for the other clustering methods. Finally, the two-step procedure clearly outperforms Clusterwise SCA-ECP in terms of retrieving the correct clustering underlying the data. We conjecture that the superior performance of the two-step procedure using ICA over Clusterwise SCA-ECP and the two-step procedure with PCA before clustering can be explained by the fact that PCA/SCA type of models do not look for independent components that are non-Gaussian. The reason for this is that the latter type of components can only be disclosed by using higher-order statistics, which ICA is relying on, whereas PCA/SCA only uses second-order statistics.
A reoccurring result in our study was that, compared to performing no ICA or PCA reduction, a (clearly) better clustering was obtained when the data were first reduced with ICA. This implies that when the cluster structure is determined by similarities and differences in FC patterns (i.e., independent and non-Gaussian components), this cluster structure cannot be recovered well by clustering the original or PCA reduced data. Indeed, in this case, ICA needs to be applied first to reveal the true clusters. An important issue when performing ICA pertains to determining an optimal number of independent components to extract. Our results show that retaining too few components has a detrimental effect on the clustering, which implies that the removed components contain relevant information to uncover the clusters. Extracting too many components, however, does not seem to harm cluster recovery (at least not for the maximal number of components tested in our study), suggesting that adding irrelevant components does not mask the cluster structure (i.e., the cluster information in the relevant components is strong enough to reveal the true clusters).
The good performance of the two-step procedure with ICA and Ward’s hierarchical clustering was also illustrated in an application of the procedure to empirical multi-subject fMRI data regarding dementia patients. In particular, the two-step procedure yielded a clustering that was stable and that corresponded to a reasonable extent with the diagnostic labels (i.e., known/given dementia subtypes). Further, the obtained cluster-specific FC patterns suggested that the salience network is more disrupted for the cluster containing mainly bvFTD patients than for the cluster consisting predominantly of AD patients, which confirms results found in previous studies. Interestingly, applying PCA (for Ward’s HC) or no data reduction (for all clustering methods) before clustering resulted in cluster solutions that captured the diagnostic labels to the same extent as ICA data reduction (for all clustering methods). It should not be forgotten that fMRI data are very noisy, even after pre-processing the data, and that the FC patterns underlying the data of both known subtypes of dementia patients may overlap to a serious extent, with only small differences in FC patterns between both subtypes being related to dementia. Indeed, in the simulation study, when the data contains large amounts of noise and clusters overlap to a substantial extent, all methods perform more or less at the same (bad) level. Finally, Clusterwise SCA-ECP obtained a clustering that did not match the diagnostic labels at all and retained cluster specific FC patterns, even after exploiting the rotational freedom of Clusterwise SCA-ECP, that did not show any relevance for dementia.
Limitations. Obviously, several limitations apply to the current study. Below, three limitations are discussed that pertain to: (1) the model selection problem, (2) the set of clustering methods adopted in this study and (3) the two-step nature of the proposed procedure.
Regarding the model selection problem, it should be noted that in the simulation study it was assumed that the true number of clusters K and—for the ICA/PCA reduction—the true number of components Q underlying the data was known. Moreover, the number of components Q underlying each patient’s fMRI data was assumed to be the same for each patient. In empirical applications, of course, the optimal number of clusters and components for a data set at hand is not known a priori and has to be determined by the researcher. Wrongly specifying the true number of components may negatively affect the retrieval of the true cluster structure underlying a given data set. It can be conjectured that an overestimation of the true number of components may retain random information that is not related to the cluster structure. Incorporating this information in the clustering phase may obscure the true cluster structure (Brusco 2004). Similarly, an underestimation of the number of components may lead to a sub-optimal cluster configuration since relevant cluster related information may be excluded during the ICA process. A re-analysis of a limited number of simulated data sets showed that under-estimation of the number of ICA components negatively affects the cluster recovery, whereas no negative effect has been encountered for overestimating Q. Future research should investigate more thoroughly how and to which extent wrongly specifying the number of components influences the cluster recovery, herewith allowing the number of components Q to differ across patients. Moreover, more work needs to be done to study the performance of several procedures to determine the optimal number of components and clusters. Especially relevant and promising in this regard is the CHull method (Ceulemans and Kiers 2006; Wilderjans et al. 2013; Vervloet et al. 2017) which simultaneously determines the optimal number of clusters and components (per patient) by finding a good balance between model fit and model complexity, with the latter being a combination of the number of components (per patient) and clusters.
A second limitation of this study pertains to the set of selected clustering methods, which is restricted to popular methods that can deal with (dis)similarities between patients as input. As is well-known, however, the type of clustering method used seriously determines the clustering result. For example, when the true clusters are non-spherical or non-convex, clustering methods like k-means fail to uncover the clusters in the data as such methods are only able to deal with spherical and convex cluster structures. Future studies therefore should investigate the nature of the patient clusters underlying multi-subject fMRI data and should compare several clustering methods in their ability to retrieve such patient clusters.
As a final limitation, the proposed procedure consists of two steps where, first, ICA components are extracted from each patient’s data and, next, clustering is performed on the basis of—the similarity between—these ICA components. A clear disadvantage of this approach is that the component extraction is performed separately from the clustering, which is referred to in the literature -and advocated against- as tandem analysis (Arabie and Hubert 1996; De Soete and Carroll 1994; Vichi and Kiers 2001; Timmerman et al. 2010; Steinley et al. 2012). As such, it is not guaranteed that the extracted ICA components contain information that allows a clear clustering of the patients. A better alternative would be to perform ICA and clustering simultaneously, which is an avenue for further study. A useful point of departure for this endeavour is similar work on the combination of clustering with Simultaneous Component Analysis (SCA; De Roover et al. 2012). In this approach, a combined model for SCA and clustering is used and the parameters of the model are estimated by means of an Alternating Least Squares (ALS) algorithm. The current study investigated the performance of Clusterwise SCA-ECP for the empirical application and a subset of the simulation design and results indicated that this method is not able to retrieve a correct clustering and/or meaningful FC patterns underlying the data. A possible reason could be that meaningful FC patterns in resting-state fMRI data are –more or less– mutually (spatially) independent and non-Gaussian distributed, which can only be disclosed using higher-order statistics (i.e., third and higher moments of the data). As Clusterwise SCA-ECP relies on second-order information (i.e., variances and covariances) solely, meaningful FC patterns may not get estimated correctly which may hamper the discovery of the true clusters underlying the data. It has to be noted that other—less constrained—variants of Clusterwise SCA exist (e.g., Clusterwise SCA-P, see De Roover et al. 2013) that may show improved results, but investigating these variants is left for future studies. In the same vein as Clusterwise SCA, a combined model for ICA and clustering could be proposed and its parameters could be estimated by ALS. A drawback, however, of such an ALS algorithm is the problem of local minima. A commonly adopted way to resolve this issue is to make use of a multi-start procedure where the algorithm is run with different (pseudo-) random and/or rational initial patient partitions (Ceulemans et al. 2007; Steinley 2003). In this regard, the two-step procedure using ICA in combination with Ward’s hierarchical clustering that was proposed in this study could be employed to identify a sensible initial -rational- patient partition. The performance of our two-step procedure as a rational start for a method that performs ICA and clustering simultaneously is a topic for further study.
Notes
It should be noted that a modified RV value can also be computed between matrices with a different number of columns. As such, the modified RV coefficient is also defined for patients that differ in the number of estimated components/FC patterns. In this paper, however, the number of components Q will always be kept equal across patients.
We will use the general term ‘objects’, which, depending on the application, refers to subjects, patients, etc.
In fMRI studies, due to computational reasons, it is common to reduce the number of voxels by enlarging the voxel size from 2 by 2 by 2 mm to 4 by 4 by 4 or 8 by 8 by 8 mm. As such, a typical brain volume (i.e., all voxels measured at one time point) contains respectively 228483, 26647 and 2553 voxels.
Note that Clusterwise SCA-P (De Roover et al. 2013), which is a less restricted version of Clusterwise SCA than Clusterwise SCA-ECP, could also be considered as a competitor for the proposed two-step clustering procedure. As this version, however, is not implemented in the Clusterwise SCA software (De Roover et al. 2012), and thus is not publicly available, we decided to not include this method in our study.
We thank the first author for kindly providing the data.
References
Agosta F, Sala S, Valsasina P, Meani A, Canu E, Magnani G, Filippi M (2013) Brain network connectivity assessed using graph theory in frontotemporal dementia. Neurology 1-10
American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders: DSM-5, 5th edn. Autor, Washington, DC
Arabie P, Hubert L (1996) Advances in cluster analysis relevant to marketing research. In: Gaul W, Pfeifer D (eds) From data to knowledge: Theoretical and practical aspects of classification, data analysis, and knowledge organization. Springer, Berlin, Heidelberg, pp 3–19
Bakeman R (2005) Recommended effect size statistics for repeated measures designs. Behav Res Methods 37(3):379–384
Banfield JD, Raftery AE (1993) Model-based Gaussian and non-Gaussian clustering. Biometrics 49:803–821
Barkhof F, Haller S, Rombouts SARB (2014) Resting-state functional MR imaging: a new window to the brain. Radiology 272(1):29–49
Beckmann CF (2012) Modelling with independent components. NeuroImage 62(2):891–901
Beckmann CF, DeLuca M, Devlin JT, Smith SM (2005) Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci 360(1457):1001–1013
Beckmann CF, Smith SM (2004) Probabilistic Independent Component Analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging 23(2):137–152
Beyer K, Goldstein J, Ramakrishnan R, Shaft U (1999) When is “nearest neighbor” meaningful? In: Beeri C, Buneman P (eds) Database theory – ICDT’99. Springer, Berlin, Heidelberg, pp 217–235
Bodenhofer U, Kothmeier A, Hochreiter S (2011) APCluster: an R package for affinity propagation clustering. Bioinformatics 27:2463–2464
Brusco MJ (2004) Clustering binary data in the presence of masking variables. Psychol Methods 9(4):510
Brusco MJ, Steinley D (2015) Affinity propagation and uncapacitated facility location problems. J Classif 32(3):443–480
Calhoun VD, Adali T, Pearlson GD, Pekar JJ (2001) A method for making group inferences from functional mri data using independent component analysis. Hum Brain Mapp 14(3):140–151. https://doi.org/10.1002/hbm.1048
Calhoun VD, Liu J, Adalı T (2009) A review of group ica for fmri data and ica for joint inference of imaging, genetic, and erp data. NeuroImage 45(1):S163–S172
Ceulemans E, Kiers HAL (2006) Selecting among three-mode principal component models of different types and complexities: a numerical convex hull based method. Br J Math Stat Psychol 59(1):133–150
Ceulemans E, Van Mechelen I, Leenen I (2007) The local minima problem in hierarchical classes analysis: an evaluation of a simulated annealing algorithm and various multistart procedures. Psychometrika 72(3):377–391
Collins FS, Varmus H (2015) A new initiative on precision medicine. N Engl J Med 372(9):793–795
Comon P (1994) Independent component analysis, a new concept? Signal Process 36:287–314
Craddock N, O’Donovan M, Owen M (2005) The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet 42(3):193–204
Cuthbert BN (2014) The rdoc framework: facilitating transition from icd/dsm to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry 13(1):28–35
Damoiseaux JS, Prater KE, Miller BL, Greicius MD (2012) Functional connectivity tracks clinical deterioration in Alzheimer’s disease. Neurobiol Aging 33(4):828-e19
De Roover K, Ceulemans E, Timmerman ME (2012) How to perform multiblock component analysis in practice. Behav Res Methods 44:41–56
Deco G, Kringelbach ML (2014) Great expectations: using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron 84(5):892–905
De Roover K, Ceulemans E, Timmerman ME, Onghena P (2013) A clusterwise simultaneous component method for capturing within-cluster differences in component variances and correlations. Br J Math Stat Psychol 66(1):81–102
De Roover K, Ceulemans E, Timmerman ME, Vansteelandt K, Stouten J, Onghena P (2012) Clusterwise simultaneous component analysis for analyzing structural differences in multivariate multiblock data. Psychol Methods 17:100–119
De Soete G, Carroll JD (1994) K-means clustering in a low-dimensional euclidean space. In: Diday E, Lechevallier Y, Schader M, Bertrand P, Burtschy B (eds) New approaches in classiffication and data analysis. Springer, Berlin, Heidelberg, pp 212–219
de Vos F, Koini M, Schouten TM, Seiler S, Grond J, Lechner A, Rombouts SARB (2018) A comprehensive analysis of resting state fmri measures to classify individual patients with Alzheimer’s disease. NeuroImage 167:62–72
Downar J, Geraci J, Salomons TV, Dunlop K, Wheeler S, McAndrews MP, Giacobbe P (2014) Anhedonia and reward-circuit connectivity distinguish nonresponders from responders to dorsomedial prefrontal repetitive transcranial magnetic stimulation in major depression. Biol Psychiatry 76(3):176–185
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Liston C (2017) Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 23(1):28
Drzezga A, Becker JA, Van Dijk KR, Sreenivasan A, Talukdar T, Sullivan C, Sperling RA (2011) Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain 134(6):1635–1646
Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD (2011) Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp 32(12):2075–2095
Esposito F, Scarabino T, Hyvarinen A, Himberg J, Formisano E, Comani S, Di Salle F (2005) Independent component analysis of fMRI group studies by self-organizing clustering. NeuroImage 25(1):193–205
Everitt BS, Landau S, Leese M, Stahl D (2011) Cluster analysis. John Wiley and Sons, New York
Fernandes BS, Williams LM, Steiner J, Leboyer M, Carvalho AF, Berk M (2017) The new field of ’precision psychiatry’. BMC Med 15(1):80
Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Mackay CE (2009) Distinct patterns of brain activity in young carriers of the apoe-e4 allele. Proc Natl Acad Sci 106(17):7209–7214
Fox MD, Raichle ME (2007) Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711
Fraley C, Raftery AE (2002) Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc 97(458):611–631
Frey BJ, Dueck D (2008) Clustering by passing messages between data points. Science 315:972–976
García V , Mollineda R A, Sánchez J S (2009). Index of balanced accuracy: a performance measure for skewed class distributions. In: Iberian conference on pattern recognition and image analysis (pp 441–448)
Golub GH, Van Loan CF (2012) Matrix computations, vol 3. JHU Press, Baltimore
Gour N, Felician O, Didic M, Koric L, Gueriot C, Chanoine V, Ranjeva JP (2014) Functional connectivity changes differ in early and late-onset Alzheimer’s disease. Hum Brain Mapp 35(7):2978–2994
Greenhouse SW, Geisser S (1959) On methods in the analysis of profile data. Psychometrika 24(2):95–112
Greicius MD (2008) Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol 21(4):424–430
Greicius MD, Srivastava G, Reiss AL, Menon V (2004) Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci 101(13):4637–4642
Hafkemeijer A, Möller C, Dopper EG, Jiskoot LC, Schouten TM, van Swieten JC et al (2015) Resting state functional connectivity differences between behavioral variant frontotemporal dementia and Alzheimer’s disease. Front Hum Neurosci 9:474
Happé F, Ronald A, Plomin R (2006) Time to give up on a single explanation for autism. Nat Neurosci 9(10):1218
Hartigan JA, Wong MA (1979) Algorithm AS 136: A K-Means clustering algorithm. J R Stat Soc Ser C (Applied Statistics) 28:100–108
Helwig N E (2015) ica: Independent Component Analysis [Computer software manual]. https://cran.r-project.org/web/packages/ica/ (R package version 1.0-1)
Hennig C (2007) Cluster-wise assessment of cluster stability. Comput Stat Data Anal 52(1):258–271
Hennig C (2018) fpc: Flexible procedures for clustering [Computer software manual]. https://CRAN.R-project.org/package=fpc (R package version 2.1-11.1)
Hubert L, Arabie P (1985) Comparing partitions. J Class 2:193–218
Hyvärinen A (1999) Fast and robust fixed-point algorithm for independent component analysis. IEEE Trans Neural Netw 10(3):626–634
Hyvärinen A, Karhunen J, Oja E (2001) Independent component analysis. John Wiley and Sons, New York
Hyvärinen A, Oja E (2000) Independent component analysis: algorithms and applications. Neural Netw 13(4–5):411–430
Indahl U G , Næs T, Liland K H (2016) A similarity index for comparing coupled matrices [Computer software manual]. https://cran.r-project.org/web/packages/MatrixCorrelation/
Insel T, Cuthbert B , Garvey M, Heinssen R, Pine D S , Quinn K, Wang P (2010) Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. American Psychiatric Association
Insel TR, Cuthbert BN (2015) Brain disorders? precisely. Science 348(6234):499–500
Jafri MJ, Pearlson GD, Stevens M, Calhoun VD (2008) A method for functional network connectivity among spatially independent resting-state components in schizophrenia. NeuroImage 39(4):1666–1681
Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM (2012) Fsl. NeuroImage 62(2):782–790
Jutten C, Herault J (1991) Blind separation of sources, Part 1: an adaptive algorithm based on neuromimetic architecture. Signal Process 24:1–10
Kaiser RH, Andrews-Hanna JR, Wager TD, Pizzagalli DA (2015) Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry 72(6):603–611
Kaufman L, Rousseeuw P (1990) Finding groups in data: an introduction to cluster analysis. Wiley, Hoboken
Kiers HAL (2000) Towards a standardized notation and terminology in multiway analysis. J Chemom 14(3):105–122
Köhn H-F, Steinley D, Brusco MJ (2010) The p-median model as a tool for clustering psychological data. Psychol Methods 15(1):87–95
Kroonenberg PM (2008) Three-mode clustering. In: Kroonenberg P (ed) Applied multiway data analysis. Wiley, Hoboken, pp 403–432
Lee T W, Lewicki M S, Sejnowski T J (1999) Unsupervised classification with non-gaussian mixture models using ica. In: Advances in neural information processing systems (pp 508–514)
Lee Y, Park B-Y, James O, Kim S-G, Park H (2017) Autism spectrum disorder related functional connectivity changes in the language network in children, adolescents and adults. Front Hum Neurosci 11:418
Li G , Guo L, Liu T (2009 May) Grouping of brain MR images via Affinity Propagation. The ... Midwest symposium on circuits and systems conference proceedings : MWSCAS. Midwest symposium on circuits and systems 2009, pp 2425-2428. http://europepmc.org/articles/PMC3011186. https://doi.org/10.1109/iscas.2009.5118290
Li Y-O, Adalı T, Calhoun VD (2007) Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 28(11):1251–1266
Liston C, Chen AC, Zebley BD, Drysdale AT, Gordon R, Leuchter B, Dubin MJ (2014) Default mode network mechanisms of transcranial magnetic stimulation in depression. Biol Psychiatry 76(7):517–526
Lorenzo-Seva U, Ten Berge JM (2006) Tucker’s congruence coefficient as a meaningful index of factor similarity. Methodology 2(2):57–64
Lynall M-E, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, Bullmore E (2010) Functional connectivity and brain networks in schizophrenia. J Neurosci 30(28):9477–9487
Maechler M , Rousseeuw P, Struyf A, Hubert M, Hornik K (2017) cluster: Cluster analysis basics and extensions [Computer software manual]. Retrieved from https://cran.r-project.org/web/packages/cluster/ (R package version 2.0.6)
Marín O (2012) Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci 13(2):107
Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K et al (2001) A probabilistic atlas and reference system for the human brain: International consortium for brain mapping (icbm). Philos Trans R Soc Lond B Biol Sci 356(1412):1293–1322
McGrath CL, Kelley ME, Holtzheimer PE, Dunlop BW, Craighead WE, Franco AR, Mayberg HS (2013) Toward a neuroimaging treatment selection biomarker for major depressive disorder. JAMA Psychiatry 70(8):821–829
Mckeown MJ, Makeig S, Brown GG, Jung T-P, Kindermann SS, Bell AJ, Sejnowski TJ (1998) Analysis of fmri data by blind separation into independent spatial components. Hum Brain Mapp 6(3):160–188
McLachlan GJ, Basford KE (1988) Mixture models: Inference and applications to clustering (Vol 84). Marcel Dekker, New York
Mezer A, Yovel Y, Pasternak O, Gorfine T, Assaf Y (2009) Cluster analysis of resting-state fmri time series. NeuroImage 45(4):1117–1125
Millan MJ, Agid Y, Brüne M, Bullmore ET, Carter CS, Clayton NS, Young LJ (2012) Cognitive dysfunction in psychiatric disorders: characteristics, causes and the quest for improved therapy. Nat Rev Drug Discov 11(2):141
Miller CH, Hamilton JP, Sacchet MD, Gotlib IH (2015) Meta-analysis of functional neuroimaging of major depressive disorder in youth. JAMA Psychiatry 72(10):1045–1053
Milligan GW, Cooper MC (1985) An examination of procedures for determining the number of clusters in a data set. Psychometrika 50(2):159–179
Milligan GW, Soon SC, Sokol LM (1983) The effect of cluster size, dimensionality, and the number of clusters on recovery of true cluster structure. IEEE Trans Pattern Anal Mach intell 5:40–47
Mojena R (1977) Hierarchical grouping methods and stopping rules: an evaluation. Comput J 20(4):359–363
Murtagh F, Legendre P (2014) Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Class 31(3):274–295
Olejnik S, Algina J (2003) Generalized eta and omega squared statistics: measures of effect size for some common research designs. Psychol Methods 8(4):434
Pannekoek JN, Veer IM, van Tol M-J, van der Werff SJ, Demenescu LR, Aleman A, van der Wee NJ (2013) Resting-state functional connectivity abnormalities in limbic and salience networks in social anxiety disorder without comorbidity. Eur Neuropsychopharmacol 23(3):186–195
Pievani M, de Haan W, Wu T, Seeley WW, Frisoni GB (2011) Functional network disruption in the degenerative dementias. Lancet Neurol 10(9):829–843
Core Team R (2017). R: A language and environment for statistical computing [Computer software manual]. Vienna, Austria. https://www.R-project.org/
Rombouts SA, Barkhof F, Goekoop R, Stam CJ, Scheltens P (2005) Altered resting state networks in mild cognitive impairment and mild alzheimer’s disease: an fmri study. Hum Brain Mapp 26(4):231–239
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Santana R, McGarry L, Bielza C, Larrañaga P, Yuste R (2013) Classification of neocortical interneurons using affinity propagation. Front Neural Circuits 7:1–13
Schacht A, Gorwood P, Boyce P, Schaffer A, Picard H (2014) Depression symptom clusters and their predictive value for treatment outcomes: results from an individual patient data meta-analysis of duloxetine trials. J Psychiatr Res 53:54–61
Schumann G, Binder EB, Holte A, de Kloet ER, Oedegaard KJ, Robbins TW et al (2014) Stratified medicine for mental disorders. Eur Neuropsychopharmacol 24(1):5–50
Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD (2009) Neurodegenerative diseases target large-scale human brain networks. Neuron 62(1):42–52
Smilde AK, Kiers HAL, Bijlsma S, Rubingh CM, van Erk MJ (2009) Matrix correlations for high-dimensional data: the modified RV-coefficient. Bioinformatics 25(3):401–405
Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Beckmann CF (2009) Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci 106(31):13040–13045
Sokal R, Michener C (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Steinley D (2003) Local optima in K-means clustering: what you don’t know may hurt you. Psychol Methods 8(3):294
Steinley D, Brusco M J (2011) K-means clustering and mixture model clustering: Reply to mclachlan (2011) and vermunt (2011)
Steinley D, Brusco MJ, Henson R (2012) Principal cluster axes: a projection pursuit index for the preservation of cluster structures in the presence of data reduction. Multivar Behav Res 47(3):463–492
Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Ser B (Statistical Methodology) 63(2):411–423
Timmerman ME, Ceulemans E, Kiers HAL, Vichi M (2010) Factorial and reduced K-means reconsidered. Comput Stat Data Anal 54(7):1858–1871
Tipping ME, Bishop CM (1999) Probabilistic principal component analysis. J R Stat Soc Ser B (Statistical Methodology) 61(3):611–622
Tokuda T, Yoshimoto J, Shimizu Y, Okada G, Takamura M, Okamoto Y, Doya K (2018) Identification of depression subtypes and relevant brain regions using a data-driven approach. Sci Rep 8(1):14082. https://doi.org/10.1038/s41598-018-32521-z
Tucker LR (1951) A method for synthesis of factor analysis studies (Personnel Research Section Rapport # 984). Department of the Army, Washington, DC
van der Laan M, Pollard K, Bryan J (2003) A new partitioning around medoids algorithm. J Stat Comput Simul 73(8):575–584
van Loo HM, de Jonge P, Romeijn J-W, Kessler RC, Schoevers RA (2012) Data-driven subtypes of major depressive disorder: a systematic review. BMC Med 10(1):156
Veer IM, Beckmann C, Van Tol M-J, Ferrarini L, Milles J, Veltman D, Rombouts SA (2010) Whole brain resting-state analysis reveals decreased functional connectivity in major depression. Front Syst Neurosci 4:41
Veer IM, Oei NY, Spinhoven P, van Buchem MA, Elzinga BM, Rombouts SA (2011) Beyond acute social stress: increased functional connectivity between amygdala and cortical midline structures. NeuroImage 57(4):1534–1541
Vervloet M, Wilderjans T F, Durieux J, Ceulemans E (2017) Multichull: A generic convex-hull-based model selection method. [Computer software manual]. https://CRAN.R-project.org/package=multichull (R package version 1.0.0)
Vichi M, Kiers HAL (2001) Factorial K-means analysis for two-way data. Comput Stat Data Anal 37(1):49–64
Viroli C (2011) Model based clustering for three-way data structures. Bayesian Anal 6(4):573–602
Ward JHJ (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58(301):236–244
Welvaert M, Durnez J, Moerkerke B, Verdoolaege G, Rosseel Y (2011) neuRosim: an R package for generating fMRI data. J Stat Softw 44(10):1–18
Weng S-J, Wiggins JL, Peltier SJ, Carrasco M, Risi S, Lord C, Monk CS (2010) Alterations of resting state functional connectivity in the default network in adolescents with autism spectrum disorders. Brain Res 1313:202–214
Wilderjans TF, Ceulemans E (2013) Clusterwise parafac to identify heterogeneity in three-way data. Chemom Intell Lab Syst 129:87–97
Wilderjans TF, Ceulemans E, Kuppens P (2012) Clusterwise HICLAS: a generic modeling strategy to trace similarities and differences in multiblock binary data. Behav Res Methods 44(2):532–545
Wilderjans TF, Ceulemans E, Meers K (2013) CHull: a generic convex-hull-based model selection method. Behav Res Methods 45(1):1–15
Wilderjans TF, Ceulemans E, Van Mechelen I (2008) The CHIC model: a global model for coupled binary data. Psychometrika 73(4):729–751
Wilderjans TF, Ceulemans E, Van Mechelen I (2012) The SIMCLAS model: simultaneous analysis of coupled binary data matrices with noise heterogeneity between and within data blocks. Psychometrika 77(4):724–740
Wilderjans TF, Depril D, Van Mechelen I (2013) Additive biclustering: a comparison of one new and two existing ALS algorithms. J Class 30(1):56–74
Zhang D, Raichle ME (2010) Disease and the brain’s dark energy. Nat Rev Neurol 6(1):15
Zhang J, Li D, Chen H, Fang F (2011) Analysis of activity in fMRI data using affinity propagation clustering. Comput Methods Biomech Biomed Eng 14(3):271–281
Acknowledgements
The research leading to the results reported in this paper was sponsored by a Talent Grant (Grant Number 406.16.563) from the Netherlands Organization for Scientific Research (NWO) awarded to Jeffrey Durieux (with as supervisors Dr. Tom F. Wilderjans and Prof. Serge A. R. B. Rombouts).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Communicated by Michel van de Velden.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Durieux, J., Wilderjans, T.F. Partitioning subjects based on high-dimensional fMRI data: comparison of several clustering methods and studying the influence of ICA data reduction in big data. Behaviormetrika 46, 271–311 (2019). https://doi.org/10.1007/s41237-019-00086-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41237-019-00086-4