Abstract
While source code of software and algorithms depicts an essential component in all fields of modern research involving data analysis and processing steps, it is uncommonly shared upon publication of results throughout disciplines. Simple guidelines to generate reproducible source code have been published. Still, code optimization supporting its repurposing to different settings is often neglected and even less thought of to be registered in catalogues for a public reuse. Though all research output should be reasonably curated in terms of reproducibility, it has been shown that researchers are frequently non-compliant with availability statements in their publications. These do not even include the use of persistent unique identifiers that would allow referencing archives of code artefacts at certain versions and time for long-lasting links to research articles. In this work, we provide an analysis on current practices of authors in open scientific journals in regard to code availability indications, FAIR principles applied to code and algorithms. We present common repositories of choice among authors. Results further show disciplinary differences of code availability in scholarly publications over the past years. We advocate proper description, archiving and referencing of source code and methods as part of the scientific knowledge, also appealing to editorial boards and reviewers for supervision.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
Sharing of digital objects produced during the course of reported research has been proposed to be essential for scientific reproducibility and integrity since over a decade [1]. Likewise, still often forgotten in the shadow of data availability options, the majority of computational science publications lack code sharing, which limits the reuse of scripts and models, and moreover, limiting reproducibility [2]. Standards for making code available are necessary in order to report all aspects of computational analyses that enable replication by other scientists [3].
Scientific articles are queried for code availability using various search engines and open data. Corresponding availability statements are inspected over time, and results differ between de facto shared code (no code/code), proper documentation (doc), persistent identifiers (PID), or possible execution (exe), such as ease of installation, provided environment, or even peer-reviewed code
The topic of reproducibility has been of concern years before 2012 [4, 5]. The term reproducibility in science has already been known around 1660 and became a matter of interest for computational sciences since everybody had access to the internet [6]. All materials and methods leading to scientific results should be available to everyone to ensure that results are reproducible; however, publishing false positives due to publication bias and other factors contributed to the reproducibility crisis [7]. Reproducibility key points include the necessity for data readability and the use of standardized dictionaries, licenses, search and citation options, machine-readable meta-data, original raw data if possible and associated processing scripts next to the final data used in analyses [8]. In general, computational code should not only be easily readable but also follow basic programming and commenting styles as well as standardized nomenclatures [9,10,11,12].
Publishers’ and funding agencies’ policies on public code availability have been suggested, as it has been shown in the past, that numerous publications on computational research had not shared the underlying developed source code [13, 14]. Likewise, institutional policies such as university research data management policies can foster corresponding best practices [15, 16]. There are some references indicating a trend towards sharing data and providing source code, but also highlighting the necessity of preservation [17, 18]. This trend may be a result of introduced policies. Data sharing requirements set by publishers can, and do in some cases, significantly effect sharing practices towards open science [19]. In this regard, it has also been shown that data sharing leads to additional scholarly citations [20]. However, simple data availability statements do not guarantee long-term availability of the indicated resources, since the lack of persistent identifiers hinder data access over time [21]. On top of that, quality of available resources is not checked in most cases, such as data or code peer review, that would contribute to trust in available online resources [22]. Additionally, non-compliance with data availability statements, such as "available upon request", has been observed [23]. On the other hand, data sharing policies by funding agencies could also influence the adoption of open science principles, though this matter is controversially discussed based on the argument that top down enforced rules are prone to constrain researchers instead of proposing alternatives, resulting in exacerbating existing practices [24].
In the past, several publishers have introduced recommendations and requirements anchored in policies and guidelines on appropriate data description, standardization, archiving and sharing of digital objects [20, 25]. Many publishers primarily provide policies and guidelines on data sharing, and only in addition, they recommend to share the underlying code of scientific findings from computational analyses, but do not provide guidelines on specific steps for code sharing. Such vague recommendations have yet to be structured and specified in detail, also in a more stringent form, in order to avoid a wide margin of interpretation.
The Transparency and Openness Promotion (TOP) guidelines define several levels for reproducibility and open science principles on digital object sharing, including code [26]. This development was driven by researchers in psychology due to the reproducibility crisis [27, 28]. Level 0 encourages code sharing or does not require authors to add a code availability statement. Level 1 requires some code availability statement. Level 2 for code deposited in a trusted repository. Level 3 adds the replication of code during the review phase of the publication next to code deposition in trusted repositories. This is partly in line with the reproducibility spectrum presented in Fig. 1. Examples for all levels translated to data sharing can be found among Wiley journals [29]. Still, there is only a introductory note that research data include methods, algorithms and software, while the term code is not mentioned. Examples for level 2 are given by Nature [30] or PLOS [31]. The latter provides specific instructions on disclosure. NeurIPS is one rare example for level 3 [32]. In exceptional domain-specific cases, guidelines for structuring code submission have been established together with code reviews during the peer review process [33].
Disciplinary differences in research practices may require domain-specific standards for disclosure of digital objects. Some research areas have broadly accepted open science as standard practice, such as for social sciences where data or code sharing practices have been used by less than half of authors in 2010 and currently this number has increased to 87% [34]. However, this does not indicate that they share code or data in each of their research works. Another study highlighted up to 18% articles to share the underlying code in case of publications on computational models [35]. In distinct disciplines, for example across medical research, data sharing rates have been shown to be consistently low at around 8% with much lower rates for public code sharing [36, 37]. A meta-analysis in the domain of medical and health sciences argues that declarations of data sharing have increased since 2016, while declared and actual public data sharing significantly differed, and code sharing prevalence did not increase, other than in the domain of computer sciences [36]. Interestingly, several factors might contribute to this effect, such as data privacy when working with data from human participants, but also cultural differences among domains.
In computational research, source code constitutes the documented specification of methods, equivalently comparable to mathematical formulas in theoretical science [38]. Thereby, in a reproducible research setting the clarity of presentation becomes a new priority. Decision making has to be documented alongside source code, while versioning systems, such as GIT, facilitate transparency [39, 40]. Software has to be documented in a instructional style, to an extent that it is described sufficiently clearly and in detail to be traceable, interpretable and reproducible. There are differences in the perception as well as presentation of data between science and technology research teams [41]. This results in a different readability of source code, since development teams start scripting by rapid prototyping and then continue to optimize for performance, whereas science teams use data and code for providing their hypotheses [38].
Readability of source code is based on comprehensibility how to understand, reuse and/or change existing source code [42]. Different levels of documentation can support this process. Unfortunately, documentation is often considered a hurdle in software development, rather than recognized as benefit. Hence, documentation issues include lack of correctness, completeness, up-to-dateness and readability [43]. Source code and its documentation should be complete, consistent and accessible [44], or high-quality clean code[45]. Provenance documentation should be made available too [46]. While working on source code documentation it is beneficial to use coding guidelines, conventions, checklists, tools such as Doxygen, as well as to write tests [47, 48]. Additionally, code readability testing has been introduced, such as pair programming and reviewing source code by others, as method to improve software quality and its documentation [49].
FAIR principles for research software have been introduced on the basis of community-endorsed principles for research software by the FAIR for Research Software working group [50]. The working group has defined research software to include source code files, algorithms, scripts, computational workflows and executables that were created during the research process, further indicating the differentiation between disciplines for software components, such as operating systems, libraries, dependencies and packages, that should be considered as software and not research software [51]. The principles can be summarized to the following high-level key messages. First, research software and associated metadata have to be easily findable for both humans and machines. The citation file format (cff) as exemplary best practice provides citation metadata for research software [52]. Secondly, it has to be retrievable via standardized protocols. Third, research software should be interoperable with other software by exchanging (meta)data through interaction via standardized application programming interfaces. Fourth, it should be both usable, in terms of that it can be executed and reusable so that it can be understood, modified, built upon, or incorporated into other software. Further details and examples for adopting the principles have been outlined in [50]. This is in line with other guidelines, such as on open research software, including the key points of downloadability, installability on different platforms, defined conditions of use, open-source code with version history and proper description of dependencies [53]. FAIR, in contrast to the term open, entails metadata such as good documentation of setup, configuration and versions [54]. In the field of computer science and related fields several guidelines have been made available how to handle software artefacts that could be further refined and integrated into discipline-specific guidelines on disclosing code along scholarly publications [55, 56]. Long-term and archivable reproducibility of analysis pipelines has been evaluated for exemplary solutions, including environment and package management [57]. These suggest several requirements, such as completeness, modular designs and scalability, minimal complexity, verifiable input and output, version control and linking descriptive narratives to code. The provision of containerized applications represents a suitable example, but also web-based and desktop applications can be processed to meet the criteria by their registration in certified or domain-specific best-practice catalogues using unique and persistent identifiers, next to the provision of a proper licence and comprehensive descriptions also on all dependencies [50].
This work, summarized in Fig. 1, investigates code availability in publications in regard to possible changes over the last decade. This longitudinal analysis on practices for publishing source code of computational studies also explores disciplinary differences and indicates several open issues on source code sharing for reproducible computational scientific publications.
2 Materials and methods
In order to search for publications based on computational scripts, we take on the example of life science research where data have to be computationally analysed, limiting the search for publications by the key term "computational model", making use of PubMed Central (PMC) which comprises millions of citations for biomedical literature from MEDLINE, life science journals and online books. Since 2018 the National Library of Medicine (NLM), the world’s largest medical library operated by the US’ federal government, released a novel functionality of PMC. This feature of associating datasets to publications is based on the aggregation of data citations and data availability statements as well as supplementary materials from secondary source databanks as ClinicalTrials.gov, GenBank, figshare, Dryad, in an "associated data" box in the page navigation bar [58].
Results of web search engines for scientific publications were tested, namely Scopus next to PMC.
The PMC search summary resulted from the following parameters: Search term "computational", and article attributes "Free full text", "Associated data”, for the years 2012–2023. Results were collected on 2024-07-08.
This search strategy was complemented with a similar query using the same search attributes without any search term restriction in order to compare results not limited to "computational".
For manual control of code availability, the number of hits in results was previously further narrowed down by using the search query "computational model" instead of "computational", collected on 2023-12-27.
The Scopus search was based on the following input query:
ALL("code availability") OR ALL("code available") OR ALL("code can be found") OR ALL("code provided") OR ALL("code is deposited"). Results were collected 2023-01-18.
For further comparison Scopus search was extended to the input query ALL("computational") in comparison with:
ALL("computational") AND (ALL("code availability") OR ALL("code available") OR ALL("code can be found") OR ALL("code provided") OR ALL("code is deposited").
Data were collected 2023-02-13. There can be slight changes in the number of results returned depending on the time of data collection because of entry numbers being continuously refined by Scopus.
Since most references on the open issue of lacking code availability originate from around 2013, and the reproducibility crisis was coined around 2012 [27], timeseries data including publication dates for the respective articles were collected in order to assess possible changes during the last years. Publications from the last decade have been investigated in the cases of search queries via Scopus and PMC. Additionally, a recently updated open dataset [59] has been utilized to compare results for code availability statements isolated from the search queries described above. This includes data from the last 5 years only.
Data were exported as or manually noted in comma-separated spreadsheets from Scopus and PMC and further processed via Python (vers. 3.8.3) available in JupyterLab (vers. 3.2.5). Statistical analysis was carried out using pandas (vers. 1.4.2) method for Pearson’s correlation. Plots were compiled using matplotlib (vers. 3.5.0), pandas (1.3.4) and seaborn (0.13.1). Output and code can be found at https://www.doi.org/10.3217/gfpp0-2vf87.
3 Results
Various forms of code availability statements can be found. These include a separate section after data availability or code availability as part of data availability. Moreover, information on code availability can be found in the methods section. And, there are other publications without any code availability statement, though authors have used forms of software artefacts for their research. Some statements refer to as "upon request", "supplementary“, "download“, "can be found“, "available in“/"available at“, or "not applicable“, instead of linking to web resources. Long-term code accessibility constitutes one important criterion to be investigated, and several code repositories were identified, such as Dryad [60], GitHub linked to Zenodo [61], Code.europa.eu [62], Code-Ocean [63], Kaggle [64], R-universe, CRAN [65], CPAN [66], PyPi [67], SourceForge [68], Software Heritage [69] and various disciplinary source code libraries, indexed, e.g. in re3data [70].
To investigate the common practice in more detail, we collected results from specified search queries as shown in the next subsections.
3.1 Specified search queries on code availability among various disciplines
Analysis of indications on code availability in scientific publications has been a manual process. We have used a specified search query in Scopus [71].
Scopus search results: all documents listed upon search query "computational" (blue), and articles only out of all documents on "computational" (orange), in comparison with documents on "computational" and the search query for code availability (green) and articles, respectively (red) (colour figure online)
Isolating code availability statements from publications by using specified search strings in Scopus are presented in Fig. 2. The isolated fraction of scientific publications holding statements on code availability in various formats highlights an increasing number of articles in the last decade. Still, the general number of publications also increases over time, presented in Fig. 3. The subfraction of articles referring to associated code correlates stronger to years than the fraction of general documents, as shown in Table 1. The category of documents comprises other types of publications, such as books and chapters, conference papers, erratums, reviews, notes and letters, next to research articles. Since many of those publications, such as books and narrative reviews, will possibly not use computational analyses, the number of articles was isolated and is presented separately.
Documents of the Scopus search on articles with associated code comprise several disciplines; the most frequent ones are highlighted in Figs. 4A, B. The results present all articles that include the search query for textual indications on potential code availability. Figure (A) on top presents the individual number of documents out of the hits for a given discipline. The most documents on code availability can be found for computer science, followed by mathematics and engineering. Disciplines of medicine and life sciences have been combined and also show an increasing number of documents over the last years. Figure (B) indicates the ratio of documents from a given discipline over the overall number of documents from the given search query on code availability for each year. In this case the ratio increases over time for the discipline of computer science. There is no significant increase of this ratio for medicine and life sciences. Since there are an increasing number of documents based on bioinformatics and data science experiments in medicine and life sciences due to the growing amount of biomedical data produced, code availability should be a major topic for this discipline. The Scopus search on code availability was not targeted disciplinary-specific. Another search web resource specific for the latter disciplines has been utilized and is presented in the next subchapter.
Scopus search results of search query for code availability: A with corresponding disciplines presented as number of documents, B ratio of discipline-specific hits in comparison with the yearly overall document number resulted from the given search query on code availability. A and B share the same legend (colour figure online)
3.2 Open dataset on code availability in publicly available articles from PubMed Central
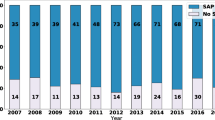
The comparator dataset [59] provided by PLOS has been reanalysed specifically for code availability statements as presented in Fig. 5. PLOS is a multidisciplinary publisher with a history to focus the fields of medicine and biology. Results are comparable to the disciplinary view in Scopus for medicine and life sciences. In case of the comparator dataset, the ratio of selected publications that share the underlying code of models or analyses has doubled since the past 5 years from 12 to almost 24%. An online location of the underlying code has been indicated in a smaller number of publications therefrom, while this number has also doubled from 10 to almost 20% between the years 2018 and 2023. Next to an indicated URL for online source code, some articles used the supplementary information for code descriptions, in many cases without presenting source scripts. The remaining subset of listed publications refer to code in the methods section without any detailed disclosure.
3.3 Specified search queries on code availability using PubMed Central
NCBI PMC provides literature from biomedical and health disciplines as well as related fields of life sciences, behavioural sciences, chemical sciences and bioengineering [72]. This was used as basic setting to search for publications on biological computational models. Free full text search results for publications with the search string "computational" in all fields are presented in Fig. 6. In addition, the NCBI filters in PMC searches allow to narrow down hits to publications with associated data. Manual inspection of hits on "computational model" indicates possibly wrong attributed associated data in case of year 2021 and years between 2013 and 2019. The data filter on "associated data" has first been introduced in 2018 retrospectively [73]. A random sample subset of publications (\(N=9\)) from those years did not include any availability statements or associated data to their experimental results. The search query using the term "computational" is further compared as ratio to all articles using resulting from no restriction in search strings, presented in Fig. 7. These lines point out the given trend of a possible wrong allocation of associated data availability to articles in all documents listed in PMC. Results from 2019, 2020, 2022 and 2023 suggest a constant number and maybe fraction of publications would include references to digital objects associated to their experimental findings. Further manual inspection of fractions in 2022 and 2023 revealed code availability statements in various formats, as part of data availability statements, separate section or part of methods. Eight and nine free full text publications from 2022 to 2023, respectively, include a code availability statement specifying a corresponding location, such as on github, figshare, OSF or others, as presented in Table 2. The table lists the publications with associated data and the repositories used for data as well as code. If mentioned in the respective manuscript, archives are also indicated. In summary for 2023 and 2022 this makes only 17 out of 31 manuscripts that provided URLs to the respective manuscript’s code. Two of the 17 URLs indicated have not been archived and were unfortunately not available anymore or at the time of analysis.
4 Discussion
Reproducibility of scientific results is essential for trust in science [74]. Scholarly research articles should therefore include all necessary descriptions as well as materials, including relevant digital objects, such as code, as suggested a decade ago. Data availability has been the primary focus of several investigations, initiatives and funding organization or publisher terms. This work intends to highlight code sharing practices in scientific publications over the last years.
Results from Scopus search query and the reanalysed comparator dataset highlight code availability statements of publications in the range of 1–24% depending on discipline. Notably, PMC search queries, given the feature of isolating publications with associated data, show inconsistent results over the last decade in the percentage of shared digital objects and therefore had to be manually inspected. The comparably low number of the corresponding publications in the years 2010 to 2012 and 2019 to 2020 as well as 2022 to 2023 points to resulting percentages between 1 and 3%. These low numbers are more realistic than the resulting high rates from the years 2013 to 2017, which have been randomly checked and identified to be falsely annotated by the search engine in several cases. This feature of associating datasets to publications provided by PMC is a valuable function for refining search queries and, however, has yet to be optimized and consequent results should be interpreted carefully and double-checked. From another point of view, the comparison of the search string "computational" to all documents listed in PMC, presented in Fig. 7, could suggest a slightly higher trend of open access documents in computational science. Still, this trend would have to be carefully reevaluated for the recent and upcoming years.
In general, both search queries in PMC and Scopus have their limitations. Next to a potential weakness of the feature for associated data, search strings used in both databases are not possible be complete: It is not possible to list all articles that, both, are based on computational analyses and include the associated published computational code.
In the cases of the Scopus query and the comparator dataset, there has been a tendency of a growing number of publications that include code availability over the last decade. Still, the disciplinary view on publications has not revealed this tendency to be true for several disciplines except for the field of computer science. This is also in line with the results from the PMC query that together with the Scopus query do not show any improvement over time towards increased code availability in life sciences and medicine. There is a disciplinary common practice to make use of certain publishers which have different policies on data or code availability in place that can range from no indication of code, to code submission and reproducibility policies, and even the provision of checklists for included source code during the article submission process [75]. Compliance to such policies would need to be checked during the peer review process.
To ensure reproducibility of published results based on computational analyses it is essential to support source code persistence and to present reusable methods. This is indeed a matter of concern, since providing URLs without archives does not suffice, since the reference needs to persist in future, ideally without access restrictions. Still, as observed, many publications providing code availability statements do not use persistent identifiers and archives for long-term accessibility of code. Corresponding examples of the manual inspection of code availability among isolated publications include several possible descriptions, such as no reference to code location, supplementary files accompanying the publication, links to github repositories, as well as common archives, such as figshare or dryad, the code-specific archive of software heritage and disciplinary curated web resources in selected cases. Notably, the open science framework (OSF) platform was indicated multiple times, showing a current trend towards adoption of open science ideals. Since data archiving has already been described in many journal policies and recommendations, these should also integrate more detailed explanatory notes on sharing code and research software.
Good quality of code documentation is another important criterion to not only help the original researchers but also other scientists in understanding and reusing the underlying scripts. Results from search queries on publications and their associated availability statements indicate that there is still plenty of room for improvement. Nevertheless, using assistive tools such as CoPilot or other code-specialized generative AI powered tools can already facilitate source code refactoring, reviewing and its documentation [76,77,78]—a step that should not be forgotten at the end of any busy day of (data) science work [47, 79]. Still, in times where systems get more complex, and highly abstracted libraries are used to work on even more complex ideas, it is essential to keep provenance in mind to be accessible as well as traceable [46].
The manual analysis of code availability statements pointed out the use of various programming languages and tools for analysis and models. Future studies could expand on this topic giving an overview on disciplinary preferences, and ease of installation, as well as provision of runtime environments and platform-independent executables, such as containerized applications. Also, the feasibility of semi-automatic approaches to such an analysis could be studied. Future research could further focus automatic generated documentation, community reviews, or more detailed statistics on preferences, such as choice of repository.
The comparison of interdisciplinary to disciplinary results on code availability among scholarly publications implicates an importance for differently elaborated recommendations and guidelines for various research fields. These could be integrated in funder and journal directives. Ultimately, semi-automatic compliance checks and adoption of policies by editorial boards and reviewers are necessary to replicate scientific results.
Data availability
Data and code can be found at https://www.doi.org/10.3217/gfpp0-2vf87.
Code availability
Data and code can be found at https://www.doi.org/10.3217/gfpp0-2vf87.
References
Hanson, B., Sugden, A., Alberts, B.: Making data maximally available. Science 331(6018), 649–649 (2011). https://doi.org/10.1126/science.1203354
Janssen, M.A., Pritchard, C., Lee, A.: On code sharing and model documentation of published individual and agent-based models. Environ. Model. Softw. 134, 104873 (2020). https://doi.org/10.1016/j.envsoft.2020.104873
Mesirov, J.P.: Accessible reproducible research. Science 327(5964), 415–416 (2010). https://doi.org/10.1126/science.1179653
Bishop, D.V.: What is the reproducibility crisis, and what can be done about it? PLoS Med. 2(8), 124 (2005)
Ioannidis, J.P.: Why most published research findings are false. PLoS Med. 2(8), 124 (2005). https://doi.org/10.1371/journal.pmed.0020124
Stodden, V.: The scientific method in practice: reproducibility in the computational sciences (2010). https://doi.org/10.2139/ssrn.1550193
Baker, M.: 1500 scientists lift the lid on reproducibility. Nature 533(7604), 452 (2016)
Tierney, N.J., Ram, K.: A realistic guide to making data available alongside code to improve reproducibility (2020). arXiv preprint arXiv:2002.11626. https://doi.org/10.48550/arXiv.2002.11626
Albertoni, R., Colantonio, S., Skrzypczyński, P., Stefanowski, J.: Reproducibility of machine learning: terminology, recommendations and open issues (2023). arXiv preprint arXiv:2302.12691. https://doi.org/10.48550/arXiv.2302.12691
Baiocchi, G.: Reproducible research in computational economics: guidelines, integrated approaches, and open source software. Comput. Econ. 30, 19–40 (2007). https://doi.org/10.1007/s10614-007-9084-4
Kernighan, B.W., Plauger, P.J.: Elements of Programming Style. McGraw-Hill Inc, New York (1974)
Kernighan, B.W.: The practice of programming. Addison-Wesley Professional (1999)
Shamir, L., Wallin, J.F., Allen, A., Berriman, B., Teuben, P., Nemiroff, R.J., Mink, J., Hanisch, R.J., DuPrie, K.: Practices in source code sharing in astrophysics. Astron. Comput. 1, 54–58 (2013). https://doi.org/10.1016/j.ascom.2013.04.001
LeVeque, R.J., Mitchell, I.M., Stodden, V.: Reproducible research for scientific computing: tools and strategies for changing the culture. Comput. Sci. Eng. 14(4), 13–17 (2012). https://doi.org/10.1109/MCSE.2012.38
Lyon, L.: The informatics transform: re-engineering libraries for the data decade. Int. J. Digit. Curation 7(1), 126–138 (2012). https://doi.org/10.2218/ijdc.v7i1.220
Reichmann, S., Klebel, T., Hasani-Mavriqi, I., Ross-Hellauer, T.: Between administration and research: understanding data management practices in an institutional context. J. Assoc. Inf. Sci. Technol. 72(11), 1415–1431 (2021). https://doi.org/10.1002/asi.24492
AlNoamany, Y., Borghi, J.A.: Towards computational reproducibility: researcher perspectives on the use and sharing of software. Peer J. Comput. Sci. 4, 163 (2018). https://doi.org/10.7717/peerj-cs.163
Deshpande, D., Sarkar, A., Guo, R., Moore, A., Darci-Maher, N., Mangul, S.: A comprehensive analysis of code and data availability in biomedical research. bioRxiv [Preprint] (2021). https://doi.org/10.31219/osf.io/uz7m5
Federer, L.M., Belter, C.W., Joubert, D.J., Livinski, A., Lu, Y.-L., Snyders, L.N., Thompson, H.: Data sharing in PLoS One: an analysis of data availability statements. PLoS ONE 13(5), 0194768 (2018). https://doi.org/10.1371/journal.pone.0194768
Christensen, G., Dafoe, A., Miguel, E., Moore, D.A., Rose, A.K.: A study of the impact of data sharing on article citations using journal policies as a natural experiment. PLoS ONE 14(12), 0225883 (2019). https://doi.org/10.1371/journal.pone.0225883
Federer, L.M.: Long-term availability of data associated with articles in plos one. PLoS ONE 17(8), 0272845 (2022). https://doi.org/10.1371/journal.pone.0272845
Kratz, J.E., Strasser, C.: Researcher perspectives on publication and peer review of data. PLoS ONE 10(2), 0117619 (2015). https://doi.org/10.1371/journal.pone.0123377
Gabelica, M., Bojčić, R., Puljak, L.: Many researchers were not compliant with their published data sharing statement: a mixed-methods study. J. Clin. Epidemiol. 150, 33–41 (2022). https://doi.org/10.1016/j.jclinepi.2022.05.019
Brysbaert, M.: The role of learned societies and grant-funding agencies in fostering a culture of open science. PsyArXiv (2021) https://doi.org/10.31234/osf.io/832me. Preprint
Mayo-Wilson, E., Grant, S., Supplee, L., Kianersi, S., Amin, A., DeHaven, A., Mellor, D.: Evaluating implementation of the transparency and openness promotion (top) guidelines: the trust process for rating journal policies, procedures, and practices. Res. Integr. Peer Rev. 6(1), 1–11 (2021). https://doi.org/10.1186/s41073-021-00112-8
Nosek, B.A., Alter, G., Banks, G.C., Borsboom, D., Bowman, S.D., Breckler, S.J., Buck, S., Chambers, C.D., Chin, G., Christensen, G.: Promoting an open research culture. Science 348(6242), 1422–1425 (2015). https://doi.org/10.1126/science.aab2374
Chiang, I.-C.A., Jhangiani, R.S., Price, P.C.: From the “Replicability Crisis” to open science practices. BCcampus, BCcampus (2015)
Nosek, B.A., Hardwicke, T.E., Moshontz, H., Allard, A., Corker, K.S., Dreber, A., Fidler, F., Hilgard, J., Kline Struhl, M., Nuijten, M.B.: Replicability, robustness, and reproducibility in psychological science. Annu. Rev. Psychol. 73, 719–748 (2022). https://doi.org/10.1146/annurev-psych-020821-114157
John Wiley & Sons, I.: Wiley’s data sharing policies. Wiley, Inc., Hoboken. Accessed 17 Jan 2024
Editorial: code sharing in the spotlight. Nat. Comput. Sci. 3(11), 907–907 (2023). https://doi.org/10.1038/s43588-023-00566-4
PLOS: Materials, Software and Code Sharing. PLOS, San Francisco, California, US. Accessed 17 Jan 2024
NeurIPS: NeurIPS 2021 code and data submission guidelines. Neural Information Processing Systems, San Diego, California, US. Accessed 17 Jan 2024
Hofner, B., Schmid, M., Edler, L.: Reproducible research in statistics: a review and guidelines for the biometrical journal. Biom. J. 58(2), 416–427 (2016). https://doi.org/10.1002/bimj.201500156
Ferguson, J., Littman, R., Christensen, G., Paluck, E.L., Swanson, N., Wang, Z., Miguel, E., Birke, D., Pezzuto, J.-H.: Survey of open science practices and attitudes in the social sciences. Nat. Commun. 14, 5401 (2023). https://doi.org/10.1038/s41467-023-41111-1
Janssen, M.A., Pritchard, C., Lee, A.: On code sharing and model documentation of published individual and agent-based models. Environ. Model. Softw. 134, 104873 (2020). https://doi.org/10.1016/j.envsoft.2020.104873
Hamilton, D.G., Hong, K., Fraser, H., Rowhani-Farid, A., Fidler, F., Page, M.J.: Prevalence and predictors of data and code sharing in the medical and health sciences: systematic review with meta-analysis of individual participant data. BMJ (2023). https://doi.org/10.1136/bmj-2023-075767
Locher, C., Le Goff, G., Le Louarn, A., Mansmann, U., Naudet, F.: Making data sharing the norm in medical research. Br. Med. J. Publ. Group (2023). https://doi.org/10.1136/bmj.p1434
Hinsen, K.: Software development for reproducible research. Comput. Sci. Eng. 15(4), 60–63 (2013). https://doi.org/10.1109/MCSE.2013.91
Ram, K.: Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 8(1), 1–8 (2013). https://doi.org/10.1186/1751-0473-8-7
Peikert, A., Brandmaier, A.M.: A reproducible data analysis workflow with r markdown, git, make, and docker. Quant. Comput. Methods Behav. Sci. (2021). https://doi.org/10.5964/qcmb.3763
Borgman, C.L., Wallis, J.C., Mayernik, M.S.: Who’s got the data? Interdependencies in science and technology collaborations. Comput. Support. Coop. Work 21, 485–523 (2012). https://doi.org/10.1007/s10606-012-9169-z
Oliveira, D., Bruno, R., Madeiral, F., Castor, F.: Evaluating code readability and legibility: An examination of human-centric studies. In: 2020 IEEE International Conference on Software Maintenance and Evolution (ICSME), pp. 348–359 (2020). https://doi.org/10.1109/ICSME46990.2020.00041 . IEEE
Aghajani, E., Nagy, C., Linares-Vásquez, M., Moreno, L., Bavota, G., Lanza, M., Shepherd, D.C.: Software documentation: the practitioners’ perspective. In: Proceedings of the ACM/IEEE 42nd International Conference on Software Engineering, pp. 590–601 (2020). https://doi.org/10.1145/3377811.3380405
Zhi, J., Garousi-Yusifoğlu, V., Sun, B., Garousi, G., Shahnewaz, S., Ruhe, G.: Cost, benefits and quality of software development documentation: a systematic mapping. J. Syst. Softw. 99, 175–198 (2015). https://doi.org/10.1016/j.jss.2014.09.042
Martin, R.C.: Clean Code: A Handbook of Agile Software Craftsmanship, 1st edn. Prentice Hall PTR, Hoboken (2008)
Kale, A., Nguyen, T., Harris, F.C., Jr., Li, C., Zhang, J., Ma, X.: Provenance documentation to enable explainable and trustworthy AI: a literature review. Data Intell. 5(1), 139–162 (2023). https://doi.org/10.1162/dint_a_00119
Hermann, S., Fehr, J.: Documenting research software in engineering science. Sci. Rep. 12(1), 6567 (2022). https://doi.org/10.1038/s41598-022-10376-9
Wang, A.Y., Wang, D., Drozdal, J., Muller, M., Park, S., Weisz, J.D., Liu, X., Wu, L., Dugan, C.: Documentation matters: human-centered AI system to assist data science code documentation in computational notebooks. ACM Trans. Comput. Hum. Interact. 29(2), 1–33 (2022). https://doi.org/10.1145/3489465
Sedano, T.: Code readability testing, an empirical study. In: 2016 IEEE 29th International Conference on Software Engineering Education and Training (CSEET), IEEE, pp. 111–117 (2016). https://doi.org/10.1109/CSEET.2016.36
Barker, M., Chue Hong, N.P., Katz, D.S., Lamprecht, A.-L., Martinez-Ortiz, C., Psomopoulos, F., Harrow, J., Castro, L.J., Gruenpeter, M., Martinez, P.A.: Introducing the fair principles for research software. Sci. Data 9(1), 622 (2022). https://doi.org/10.1038/s41597-022-01710-x
Chue Hong, N.P., Katz, D.S., Barker, M., Lamprecht, A.-L., Martinez, C., Psomopoulos, F.E., Harrow, J., Castro, L.J., Gruenpeter, M., Martinez, P.A., Honeyman, T., Struck, A., Lee, A., Loewe, A., Werkhoven, B., Jones, C., Garijo, D., Plomp, E., Genova, F., Shanahan, H., Leng, J., Hellström, M., Sandström, M., Sinha, M., Kuzak, M., Herterich, P., Zhang, Q., Islam, S., Sansone, S.-A., Pollard, T., Atmojo, U.D., Williams, A., Czerniak, A., Niehues, A., Fouilloux, A.C., Desinghu, B., Goble, C., Richard, C., Gray, C., Erdmann, C., Nüst, D., Tartarini, D., Ranguelova, E., Anzt, H., Todorov, I., McNally, J., Moldon, J., Burnett, J., Garrido-Sánchez, J., Belhajjame, K., Sesink, L., Hwang, L., Tovani-Palone, M.R., Wilkinson, M.D., Servillat, M., Liffers, M., Fox, M., Miljković, N., Lynch, N., Martinez Lavanchy, P., Gesing, S., Stevens, S., Martinez Cuesta, S., Peroni, S., Soiland-Reyes, S., Bakker, T., Rabemanantsoa, T., Sochat, V., Yehudi, Y., WG, R.F.: FAIR Principles for Research Software (FAIR4RS Principles). Zenodo (2022) https://doi.org/10.15497/RDA00068
Druskat, S.: The citation file format: providing citation metadata for research software. In: deRSE23 - Conference for Research Software Engineering in Germany. Zenodo, Paderborn, Germany (2023). https://doi.org/10.5281/zenodo.7655140
Bezjak, S., Clyburne-Sherin, A., Conzett, P., Fernandes, P.L., Görögh, E., Helbig, K., Kramer, B., Labastida, I., Niemeyer, K., Psomopoulos, F., Ross-Hellauer, T., et al.: Open Science Training Handbook. Zenodo. computer software (2018). https://doi.org/10.5281/zenodo.1212495. (https://open-science-training-handbook.gitbook.io/book/02opensciencebasics/03openresearchsoftwareandopensource)
Tennant, J.P., Agrawal, R., Baždarić, K., Brassard, D., Crick, T., Dunleavy, D.J., Rhys Evans, T., Gardner, N., Gonzalez-Marquez, M., Graziotin, D., et al.: A tale of two’opens’: intersections between free and open source software and open scholarship (2020)
Heumüller, R., Nielebock, S., Krüger, J., Ortmeier, F.: Publish or perish, but do not forget your software artifacts. Empir. Soft. Eng. 25(6), 4585–4616 (2020). https://doi.org/10.1007/s10664-020-09851-6
Wilson, G., Bryan, J., Cranston, K., Kitzes, J., Nederbragt, L., Teal, T.K.: Good enough practices in scientific computing. PLOS Comput. Biol. 13(6), 1–20 (2017). https://doi.org/10.1371/journal.pcbi.1005510
Akhlaghi, M., Infante-Sainz, R., Roukema, B.F., Khellat, M., Valls-Gabaud, D., Baena-Gallé, R.: Toward long-term and archivable reproducibility. Comput. Sci. Eng. 23(3), 82–91 (2021). https://doi.org/10.1109/MCSE.2021.3072860
National Library of Medicine: Associated data in pmc. NLM Tech. Bull. 425, b4 (2018)
L Cadwallader, I.H.L.: PLOS Open Science Indicators—Comparator-Dataset_v5_Dec23.csv. Figshare. Last update: 02-11-2023 (2022). https://doi.org/10.6084/m9.figshare.21687686
Vision, T.: The dryad digital repository: published evolutionary data as part of the greater data ecosystem. Nat. Preced. (2010). https://doi.org/10.1038/npre.2010.4595.1
Troupin, C., Muñoz, C., Fernández, J.G., Rújula, M.À.: Scientific results traceability: software citation using github and zenodo. In: IMDIS 2018 International Conference on Marine Data and Information Systems (2018). https://imdis.seadatanet.org/content/download/122158/file/IMDIS_2018_submission_66.pdf
The European Commission: COMMISSION DECISION of 8 December 2021 on the open source licensing and reuse of Commission software (2021/C 495 I/01). https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32021D1209(01) (2021)
Cheifet, B.: Promoting reproducibility with code ocean. Genome Biol. 22(1), 1–2 (2021). https://doi.org/10.1186/s13059-021-02299-x
Banachewicz, K., Massaron, L., Goldbloom, A.: The Kaggle Book: Data Analysis and Machine Learning for Competitive Data Science. Packt Publishing Ltd (2022). ISBN: 978-1-80181-747-9
Hornik, K.: The comprehensive r archive network. Wiley Interdiscip. Rev. Comput. Stat. 4(4), 394–398 (2012). https://doi.org/10.1002/wics.1212
Tregar, S.: Cpan. In: Writing Perl Modules for CPAN, pp. 1–20. Springer, Berlin (2002). https://doi.org/10.1007/978-1-4302-1152-5_1
Bommarito, E., Bommarito, M.J.: An empirical analysis of the python package index (pypi). Available at SSRN 3426281 (2019). https://doi.org/10.2139/ssrn.3426281
Van Antwerp, M., Madey, G.: Advances in the sourceforge research data archive. In: Workshop on Public Data About Software Development (WoPDaSD) at The 4th International Conference on Open Source Systems, Milan, Italy, pp. 1–6 (2008). Citeseer. https://citeseerx.ist.psu.edu/pdf/9413345684378a5c5cbe6cf8e43201112c8716c2
Di Cosmo, R., Zacchiroli, S.: Software heritage: Why and how to preserve software source code. In: iPRES 2017-14th International Conference on Digital Preservation, pp. 1–10 (2017). https://hal.science/hal-01590958
Pampel, H., Vierkant, P., Scholze, F., Bertelmann, R., Kindling, M., Klump, J., Goebelbecker, H.-J., Gundlach, J., Schirmbacher, P., Dierolf, U.: Making research data repositories visible: the re3data.org registry. PLoS ONE 8(11), 78080 (2013). https://doi.org/10.1371/journal.pone.0078080
Burnham, J.F.: Scopus database: a review. Biomed. Digit. Libr. 3(1), 1–8 (2006). https://doi.org/10.1186/1742-5581-3-1
NIH, N.L.o.M.: About—PubMed. https://pubmed.ncbi.nlm.nih.gov/about/. Last update: August 15, 2023. Accessed 13 July 2024 (2023)
Bulletin, N.T.: Data filters in PMC and pubmed. NLM Technical Bulletin. 2018 Mar-Apr. National Institutes of Health. Last update: April 24 2018. Accessed 13 July 2024. (2018). https://www.nlm.nih.gov/pubs/techbull/ma18/brief/ma18_pmc_data_filters.html
Willis, C., Stodden, V.: Trust but verify: how to leverage policies, workflows, and infrastructure to ensure computational reproducibility in publication. Harv. Data Sci. Rev. (2020). https://doi.org/10.1162/99608f92.25982dcf
Walters, W.P.: Code sharing in the open science era. J. Chem. Inf. Model. 60(10), 4417–4420 (2020). https://doi.org/10.1021/acs.jcim.0c01000
Ebert, C., Louridas, P.: Generative AI for software practitioners. IEEE Softw. 40(4), 30–38 (2023). https://doi.org/10.1109/MS.2023.3265877
Moradi Dakhel, A., Majdinasab, V., Nikanjam, A., Khomh, F., Desmarais, M.C., Jiang, Z.M.J.: Github copilot AI pair programmer: asset or liability? J. Syst. Softw. 203, 111734 (2023). https://doi.org/10.1016/j.jss.2023.111734
Hadi, M.U., Qureshi, R., Shah, A., Irfan, M., Zafar, A., Shaikh, M.B., Akhtar, N., Wu, J., Mirjalili, S.: A survey on large language models: Applications, challenges, limitations, and practical usage. Authorea Preprints (2023). https://doi.org/10.36227/techrxiv.23589741.v1
Trisovic, A., Lau, M.K., Pasquier, T., Crosas, M.: A large-scale study on research code quality and execution. Sci. Data 9(1), 60 (2022). https://doi.org/10.1038/s41597-022-01143-6
Acknowledgements
We thank all open data providers and scientists pursuing best practices for open science. We thank Iain Hrynaszkiewicz from the Public Library of Science making us aware of open data from analyses of data availability among selected publications.
Funding
Open access funding provided by Graz University of Technology. This research received no external funding.
Author information
Authors and Affiliations
Contributions
C.J. helped in conceptualization, methodology, investigation, data curation; F.J. was involved in formal analysis; I.H. helped in resources; C.J. and F.J. contributed to writing—original draft preparation and visualization; S.S., J.S. and B.S. helped in writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Consent for publication
All authors approved the manuscript and give their consent for submission and publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jean-Quartier, C., Jeanquartier, F., Stryeck, S. et al. Sharing practices of software artefacts and source code for reproducible research. Int J Data Sci Anal (2024). https://doi.org/10.1007/s41060-024-00617-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41060-024-00617-7