Abstract
Non-tuberculous mycobacterium (NTM) infections are often clinically challenging, with lengthy antibiotic regimens that fail to resolve the infections with few good outcomes remaining. Mycobacteriophages—viruses that infect Mycobacterium hosts—show promise as therapeutic agents for NTM infections and have been used in 20 compassionate use cases. Favorable outcomes were observed in many but not all cases, although the phages show exceptional safety profiles and no evidence of phage resistance was observed, even when only a single phage was administered. Phage-specific antibodies are commonly present following intravenous administration and are often neutralizing for the phage in vitro. However, phage neutralization does not consistently correlate with poor treatment outcomes and may not be a therapeutic limitation in all patients, even when immunocompetent. Currently, the therapeutic potential of phages is substantially limited by the great variation in phage susceptibility and a relatively small repertoire of therapeutically useful phages. As many as 45% of clinical isolates can have a smooth colony morphotype, and phages that both efficiently infect and kill these strains have yet to be described. In contrast, ~ 75% of rough strains are susceptible to and killed by one or more phages and therapeutic options can be considered on a compassionate use basis. Although therapies must currently be personalized, elucidating the determinants of phage host specificity, expanding the useful phage repertoire, and identifying the key determinants of clinical outcomes will reveal their full therapeutic potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Bacteriophages show promise for treating NTM infections. |
Variation among NTM clinical isolates currently demands personalized treatment. |
Phage treatment of NTM infections is not evidently limited by phage resistance. |
Antibody neutralization does not obviously correlate with patient outcomes. |
Introduction
The therapeutic use of bacteriophages has been contemplated for nearly 100 years, nearly as long they have been recognized as viruses that infect bacterial hosts [1]. At the time of the first descriptions of phages in the latter part of the First World War, the age of small molecule antibiotics had not even been imagined. Infectious diseases were a major contribution to human mortality and there was naturally great interest in the potential of these new marvels in medicine [2]. There were many early claims of success and phage therapy became widespread in Eastern Europe and the Soviet Union [3]. The first antibiotics—penicillin and streptomycin—became more widely available in the early 1940s and came into widespread use in the Western world. Phage therapy continued for many years in Eastern Europe, and there are active therapy programs in Georgia and Poland to this day [4, 5]. There was little interest in phage therapies in Europe and the US because of the great effectiveness of antibiotics, but this view has changed with the predictable and inevitable development of antibiotic resistance in bacterial pathogens [6].
The threat of antibiotic resistance has grown to an alarming level, with predictions of such infections rising to pandemic level within only a few years [7]. Additionally, there are several important pathogens that are intrinsically resistant to antibiotics; and despite evidence of sensitivity in vitro, antibiotic use is not guaranteed to lead to clearance of the infection [8, 9]. These collective concerns have stirred renewed interest in the therapeutic use of phages and although enthusiasm has waxed and waned over the past 30 years or so, today there is hope that the potential may finally be fully realized [6, 10].
Mycobacteria are some of nastiest pathogens of Man. Tuberculosis, caused by infection with Mycobacterium tuberculosis, remains widespread throughout the developing world, and highly drug resistant strains are common. Nontuberculous mycobacteria (NTM)—including Mycobacterium abscessus, Mycobacterium avium among others—are generally opportunistic pathogens and are prevalent in patients with cystic fibrosis and those immunocompromised through HIV infection or otherwise immunosuppressed for a variety of reasons. They are typically intrinsically antibiotic resistant but can also have acquired antibiotic resistances [8]. NTM infections are more prominent than M. tuberculosis infections in most of the developed world and are unwelcome news for both the patient and the physician who recognizes the need to embark on lengthy and often failing drug regimens with harmful side effects [11, 12]. This then raises the question as to whether phages have any therapeutic role to play in treatment of pulmonary NTM infections. Several recent reviews may provide helpful complementary perspectives [6, 13,14,15,16,17,18,19,20].
The First Therapeutic Use of Phages for Treatment of an NTM Infection
In 2017, two pediatric patients in London faced similar challenges: both were persons with cystic fibrosis, both had received bilateral lung transplants together with immunosuppression to support the new lungs, and both had drug resistant disseminated M. abscessus infections [21]. Both were referred for palliative care, and physicians had exhausted all treatment options to control the NTM disease. Prompted by the parents of one of the patients who had read about phage therapy, the physicians reached out to learn if phages might offer some hope. However, there were no reported cases of phage therapy for NTM infections, and this meant starting near ground zero.
Although several laboratories have explored isolating phages on M. abscessus strains, few if any had been reported at that time. However, a very large collection of phages had been amassed and archived at the University of Pittsburgh that infect the related strain Mycobacterium smegmatis (discussed in further detail below). M. smegmatis and M. abscessus are more like distant cousins than siblings, so it was likely that most of these phages would not infect M. abscessus. However, prior studies showed that at least some can efficiently infect M. tuberculosis, another distant cousin [22, 23]. So, with detailed knowledge of the phage genomes and some insights into host range, a subset of about 200 phages were screened for their ability to infect the two M. abscessus strains from London [21]. The pickings were slim. For one strain (designated GD02), no potentially useful phages were identified. For the other strain (GD01) three candidates were found, but only one (phage Muddy) efficiently killed the strain, and further work was needed [21] (Fig. 1). Two of the three phages were temperate (BPs and ZoeJ), and lysogenic survivors arise at high frequency (Fig. 2A) [24, 25]. Engineering tools had been developed that enabled removal of all or part of the repressor genes of these [26, 27], thus converting them from being temperate to obligatorily lytic and efficient killers of GD01 (Fig. 2B). One of these (phage BPs; Fig. 1) infects GD01 at only low frequency, but a host range mutant (HRM) was isolated that overcomes the host barrier and now infects GD01 as efficiently as it infects and kills M. smegmatis [21].
Mycobacteriophages BPs and Muddy. Mycobacteriophages BPs and Muddy have both been used for therapeutic treatment of NTM patients. A Electron micrographs show that BPs and Muddy are morphologically similar with icosahedral capsids containing dsDNA, and long flexible non-contractile tails; all other mycobacteriophages used therapeutically to-date share this overall morphology. Scale bars are 100 nm. B Although morphologically similar, BPs and Muddy are genomically distinct. Genome maps of Muddy and BPs have been aligned to illustrate that there is only very limited nucleotide sequence similarity (spectrum-colored shading between genomes, with red being least similar above a threshold BLASTN E-value of 10–4). Muddy and BPs are assigned as members of Cluster AB and G1, as indicated. The organization of the genes (colored boxes) expressed early and late are shown with arrows reflecting the direction of transcription; descriptions over the arrows reflect some gene functions, although the functions of many genes are not known. Muddy is a lytic phage and does not lysogenize (see Fig. 2), whereas BPs is a temperate phage and has genes coding for an integrase and repressor (‘Lysogeny’), as shown. The Muddy and BPs genomes are 48,228 bp and 41,901 bp long, and code for 71 and 63 genes, respectively. Only seven of these genes are homologues and shared by both phages (indicted by black lines with dumbbells); all are virion structural proteins
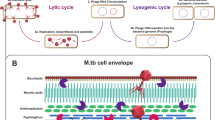
Bacteriophage life cycles and phage engineering. A Many phages can be classified as being either ‘lytic’ or ‘temperate’. Lytic phages typically infect the host bacterium through adsorption, DNA injection, DNA replication, assembly of progeny phage particles, and lysis of the bacterial cell. The only outcome is phage replication and cell death due to lysis. This is illustrated by the clear plaques of a lytic phage as seen by infection of serial dilutions of a phage stock on solid medium (top). The spots appear dark, compared to the surrounding lawn of cells because all of the cells in the spots are killed as a result of phage lysis. Temperate phages similarly adsorb to the host bacterium and inject their DNA. Most of these infections result in lytic growth as described above, although a subset (the proportion varies greatly depending on the conditions) enter a state of lysogeny, in which the phage DNA integrates into the bacterial genome to form a prophage, and the lytic phage functions are switched off. This behavior is illustrated by serial dilutions of a temperate phage stock on solid medium, as shown at the bottom. Turbid plaques are formed in which bacterial lysogens are growing. The path followed is influenced by many factors including the multiplicity of infection (MOI) and nutrient availability. Lysogens can be readily recovered from turbid plaques, purified, and stably propagated. However, the culture medium usually contains phage particles produced from low-level spontaneous induction of the prophage. Image made using BioRender. B Phage genome engineering. The green-shaded box illustrates the use of the Bacteriophage Recombineering of Electroporated DNA (BRED) strategy [26] to delete a hypothetical gene such as gene 43. A synthetic DNA substrate containing sequences flanking gene 43 is co-electroporated with phage genomic DNA (gDNA) into a recombineering strain (e.g., M. smegmatis mc2155pJV138), and plaques recovered on solid medium. Individual plaques are recovered (1° plaques), screened by PCR for the presence of the mutation, plaque-purified and rescreened (2° plaques) to find a homogenous deletion mutation. In the CRISPY-BRED adaptation [27], first (aqua-shaded box) a sgRNA-expressing plasmid targeting gene 43 is constructed and demonstrated to reduce phage infection when sgRNA expression is induced with ATc (+ ATc). Then the electroporated cells (from green-shaded box) are plated on solid medium with the sgRNA-expressing cells, with ATc. This facilitates counter-selection against the wild-type phage, and efficient recovery of the desired mutants. Parts A and B adapted from Hatfull et al. [6] and Wetzel et al. [27] with permissions
The three phages were amplified to high titer, purified extensively, and the preparations shown to be sterile and free of endotoxins [21]. It was decided to use all three phages as a cocktail to reduce the likelihood of the strain becoming resistant to the phages, and as the patient was pharmaceutically immunosuppressed a neutralizing immune response was not anticipated. After satisfying a vigorous regulatory review, the phages were shipped to London and therapy was initiated about 6 months after the strains had been shipped to Pittsburgh. The phages were administered twice daily intravenously at a dose of 109 plaque-forming units (PFU), with escalating dosage over the first hour of infusion [21]. No serious adverse reactions were reported, but treatment was anticipated to be lengthy, similar to antibiotic treatments for NTM infections [21]. After 6 weeks, substantial clinical improvement was observed, with resolution of infection in the liver, reduction of many but not all of the extensive skin nodules, and closure of the infected sternal transplant wound [21]. She continued to improve and enjoy life, although subsequently succumbed about three and a half years after the start of treatment to complications unrelated to the phage therapy. Overall, the therapy succeeded in diminishing the infection, although it was likely not completely cleared. However, no resistance to the phages was observed, and the phages performed better than the long and complex series of antibiotic regimens [21]. It offered sufficient encouragement to explore additional therapies.
The Origins and Benefits of a Large Phage Collection
Progress in this first case was facilitated by the availability of a large and diverse phage collection, because identifying potentially therapeutic phages was challenging. Where did the collection come from and what have we learned from it? Although some mycobacteriophages were described more than seventy years ago—many in the context of typing clinical isolates of M. tuberculosis—the first complete genome sequence was reported in 1993 [28]. Three more genome sequences were reported over the next 7 years, but comparative analysis indicated that many more would need to be isolated and sequenced to provide a fuller picture of phage diversity, evolution, and origins. In 2002 the Phage Hunters Integrating Research and Education (PHIRE) program was developed to provide novice student scientists (undergraduate and high school students) with an experience in scientific discovery, while also expanding the number of sequenced mycobacteriophage genomes [29]. In 2008, the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program was started which extended this platform to multiple institutions and targeted first year undergraduate students [30, 31]. By using a common programmatic infrastructure, phage discovery and genomics could be implemented at very large scale, with over 40,000 students through the program to-date, and over 20,000 phages isolated; over 4000 have been sequenced and fully annotated [31].
These phages have been isolated on a variety of bacterial strains within the phylum Actinobacteria, with about 10,000 isolated on M. smegmatis mc2155, the largest set of phages isolated on a single common bacterial strain; these are described at https://phagesdb.org [32]. Over 2,000 of these have been sequenced, providing a high-resolution view of their diversity [15]. Overall, there appears to be an underlying continuum of diversity, although the collection is heterogenous, so some types are much more prevalent than others [33]. It has been pragmatically useful to assign related phages to groups designated as clusters (e.g., Cluster A, B, C etc.). Many of these have apparent divisions enabling further grouping into subclusters (e.g., Subcluster A1, A2, A3 etc.); phages with no close relatives are designated as singletons [16]. Currently, there are 31 clusters and seven singletons. The parameters for cluster designation have evolved somewhat as the number of phages has grown, and currently phages within a cluster typically share at least 35% of their genes [34]. The overall heterogeneity is reflected in the very large number of phages in some clusters (e.g., > 700 and > 350 in Clusters A and B, respectively), while the seven singletons currently have no close relatives.
This genomic information is extremely valuable in the context of advancing therapeutic utility, as there is a relationship between genome similarity and host range. This is reflected in experiments to determine which subset of phages infect other strains of M. smegmatis (Jucho or KMD8), or M. tuberculosis [22]. Most, but not all, of the phages infect the other two M. smegmatis strains, but some do not and there is a statistically significant correlation between this and their designated cluster. For the more distantly related M. tuberculosis, the correlation is much more robust, and only phages within a few clusters/subclusters infect M. tuberculosis H37Rv. Specifically, these are Subclusters A2 and A3, Cluster G, Cluster K, and Cluster AB; but it should be noted that there is considerable diversity within clusters/subclusters, and not all phages within a group necessarily infect M. tuberculosis [23].
The search for phages that infect M. abscessus GD01 suggested that the phages that infect M. tuberculosis have generally broad host ranges, and phages in those clusters/subclusters are more likely to infect M. abscessus. Thus, the phages used in the initial therapy—BPs, ZoeJ, and Muddy—belong to Clusters G, K, and AB, respectively. Nonetheless, it was also apparent that not all phages in these groups infect GD01, and none of them efficiently infected the other tested strain, M. abscessus GD02.
M. abscessus Strains Vary Greatly in their Phage Infection Profiles
With news of the favorable outcomes in the first NTM phage therapy case, physicians have inquired about the possibility of using phages to treat other patients with similar NTM infections [35]. These requests have originated globally and at a rate of one every 2–3 days, the majority being cystic fibrosis patients and the most common pathogen being M. abscessus. Clinical isolates are typically sent to the University of Pittsburgh for analysis and are screened for phage susceptibility using a standard plaque assay (Fig. 3A); a panel of 24–36 phages is typically used that contains the best therapeutic phage candidates. Some strains show similar efficiencies of plaquing (EOP) on the clinical isolate as the M. smegmatis control strain, whereas others may not be efficiently infected by any of the tested phages (Fig. 3B). For the latter cases, therapies are not available, and any further exploration would require a much more detailed investigation, which is not usually feasible. However, if one or more phages efficiently infect the strain, then further analyses are done to determine whether the phage also efficiently kills the strain when it is challenged in vitro (Fig. 3C). Although most strains are killed well, a subset of strains are not efficiently killed, even though the lytic phage infects with an EOP of near one (Fig. 3C). Overall, these clinical isolates represent a major resource of diverse strains for elucidating the phage infection profiles, and what determines them. Several important findings emerge.
Screening clinical isolates for phage sensitivity and killing. A On receipt of a patient isolate (usually on a slant), the strain is streaked on solid growth medium and the colony morphotype (rough or smooth) is determined; the slant is also inoculated into liquid culture. If the sample is homogenous, the liquid culture can be tested, but if not, liquid cultures are grown from single colonies. These are then tested for phage sensitivity using a standard plaque assay and a collection of candidate phages, in which phage lysates are tenfold serially diluted and the efficiency of plaquing (EOP) on the clinical isolate is compared with a control strain (usually M. smegmatis). DNA is also prepared for DNA sequencing. If phages are identified that efficiently infect the clinical isolate (typically EOP > 0.1), then the effectiveness of killing the strain is evaluated in a ‘killing assay’ using a range of bacterial and phage concentrations. A ‘survival assay’ is also performed in which ~ 2 × 108 cells are incubated in liquid culture with phages at a multiplicity of infection (MOI) of 10, and then plated on solid media to determine how many cells survive. For M. abscessus, this process takes 3–4 weeks. B Examples of phage screening assays. On the left (‘Green light’) is a positive outcome in which many phages (numbered 1–11) efficiently infect the M. abscessus strains (designated generically as GDxx), and screening can proceed to the killing and survival assays. On the right is shown a poor outcome (’Red light’) in which none of the phages tested efficiently infect the strain and no further testing is done. C Not all phages that infect efficiently (i.e., EOP > 0.1), also kill efficiently. On left is shown the survival assay using a phage that both efficiently infects and kills the strain. The strain was incubated either with or without the phage, and then plated on solid medium. Note that the cells are not only killed well, but there are no phage-resistant colonies growing on the plate, suggesting that resistance only occurs at very low frequencies. On the right is an example of a strain that is efficiently infected by a phage (EOP > 0.1) but not killed efficiently, and there is abundant growth from the phage-infected cells
First, there are relatively few (genomically distinct) M. smegmatis phages that infect any M. abscessus strain [36], and only seven have been used therapeutically (Table 1) [35]. The testing of these for infection of an initial series of 82 M. abscessus clinical isolates revealed the considerable variation in the phage infection profiles, even among strains that are very closely related (Fig. 4A). These variations were apparent in comparing the efficiencies of plaque formation (efficiency of plaquing or EOP) between the control strain (M. smegmatis mc2155) and each M. abscessus isolate. Second, there is a profound difference between the behavior of smooth and rough colony morphotype strains (Fig. 4B). Of the 82 strains tested, about 45% have a smooth colony morphotype associated with environmental isolates with abundant glycopeptidolipids (GPLs) in their cell walls [36, 37]. The other 55% have a rough colony morphotype and mutations in the GPL biosynthetic genes [36]. Phages were identified that efficiently infect some of the smooth strains, but they are not efficiently killed by these phages, even though the phages are lytic on other strains (Fig. 4C). As a consequence, no therapeutically useful phages have yet been identified for any of the smooth strains. This phenotype is also observed for some rough strains, but it is relatively rare [35].
Variations in phage sensitivity of M. abscessus clinical isolates. A The phylogenies of clinical isolates of M. abscessus subsp. massiliense (designated GDxx) determined by their shared sequences is shown with longer branch lengths reflecting more distant relationships. Each strain is designated as having either a rough (R) or smooth (S) colony morphotype. The infection by individual phages (or none) of each strain at an EOP > 0.1 relative to M. smegmatis is shown as a filled circle. If the phage infects well but does not kill efficiently, an X is superimposed on the circle. B Examples of strains with rough and smooth colony morphotypes are shown. C Phage infection profiles of a smooth strain (GD84, top) and a rough strain (GD100B, bottom). For the smooth strain, Itos infects the strains efficiently, but does not kill it efficiently as illustrated to the right. The rough strain is efficiently infected by both of the BPs derivatives, which also efficiently kill the strain, as shown in the survival assay on the right. Adapted from Dedrick et al. [36] with permission
The key finding of this analysis of phage host range and strain variation is that any potential therapy needs to be personalized to the patient and the specific strain, at least until we have a fuller understanding of the determinants of host range and find ways to construct phage cocktails that more broadly infect and kill M. abscessus clinical isolates.
Does Phage Therapy for Pulmonary NTM Infections Work?
The screening of NTM isolates for phage susceptibilities showed that at least one potentially therapeutically useful phage was identified for about 75% of the rough colony morphotype isolates [36]. A subset of these satisfying clinical and regulatory parameters were able to progress to therapeutic applications on a compassionate use basis, and a consecutive series of 20 case studies has been reported [35]. A summary of these therapies and their outcomes is shown in Table 2. Although each is essentially an anecdotal report—and most patients have multiple clinical complications—some general indications can be drawn. First, no serious adverse events were reported, and the safety profile appears to be very good while administering a twice-daily dosage—at 109 plaque-forming units (PFU)/dose. Secondly, favorable microbiological outcomes were reported in eleven cases, with four showing no evident improvement; five cases were not easily interpretable. Although this suggests that phages are no magic bullet for treating NTM infections, some patients appeared to benefit from the intervention. Because these patients had exhausted available antibiotic options, the outcomes suggest that further investigations are warranted. Thirdly, there was no evidence of resistance to the phages emerging, even in the eleven cases where only a single phage was used, because only one was available. This is somewhat unexpected, although phage resistance in M. abscessus only occurs at low frequencies in vitro [36]. There was also no evidence of other phenotypic changes in the pathogens after initiation of phage administration, or resident strains had been replaced by different strains, resulting in treatment failure [35]. Lastly, antibody responses were observed in most patients given an IV administration which for many phages were neutralizing in vitro. However, how this relates to clinical outcomes is unclear. In one case, a decline in efficacy correlated with antibody neutralization [35], but in other patients antibody neutralization didn’t prevent favorable outcomes [35, 38, 39]. We also note that as all of these cases were on a compassionate use basis, all patients continued on their extant antibiotic regimens, and specific phage and antibiotic combinations could give therapeutic activity, as noted for a prior case [40]. Clearly further studies are warranted.
In summary, these compassionate use cases, all with FDA eIND approval—or equivalent in other countries—have provided substantial insights that are invaluable for designing and implementing clinical trials. Studies looking at various biomarkers during therapy are especially helpful [35, 38]. However, designing clinical trials is confounded by the strain variation in phage infection. Excluding patients with smooth morphotype strains would be feasible, but even the best possible phage cocktail would only attack about 75% of the rough strains. And because of the long treatment regiments (many months) antibody neutralization could warrant a phased use of the phages rather than combining into a single cocktail with only one shot at therapy. It is a high priority to explore how to expand the phage repertoire to kill a higher proportion of rough strains, and to figure a solution to the challenges with smooth strains.
Strategies for Expanding Phage Therapeutic Utility
Two approaches have been useful in expanding the phage repertoire and the spectrum of strains that are infected and may be useful for further development. First, many of the mycobacteriophages are temperate (Fig. 2), and therefore need to be engineered before they are therapeutically useful, as described for the initial screen against M. abscessus GD01. The simple conversion from temperate to lytic behavior can be achieved by removal of all or part of the phage repressor gene (Fig. 2). Arguably it is best to also delete the integrase gene. The rationale for this is based on observations made testing for phage infection of M. tuberculosis [23]. When challenged with a derivative in which only the repressor was removed, phage resistant survivors were recovered in which parts of the phage genome were integrated into the host genome and presumably expressed genes conferring resistance through surface exclusion [23]. Removing the integrase gene should minimize resistance occurring through this mechanism. In general, the methods for engineering these phage genomes are well established, and use a recombineering system that increases the frequency of recombination in M. smegmatis [41] and is well adapted for phage manipulations (Fig. 2B) [26, 42]. The overall efficiency of this engineering can be enhanced by using CRISPR-mediated counter-selection [27].
Second is the use of phage evolution to obtain derivatives with broader host infectivity. This was used to make phage BPs utile for the first therapy case discussed above [21], but another example is the evolution of phage D29, which itself does not efficiently infect any M. abscessus strain [36]. However, host range mutants (HRMs) could be readily isolated on strain M. abscessus GD40, which now efficiently infect this strain when purified and characterized [36]. This HRM also infects some other M. abscessus strains and was deployed therapeutically to treat two other patients infected with different strains that are sensitive to this phage variant [35, 38]. It is likely that there are other phages which could be mutationally ‘activated’ likewise.
Another promising direction to expand the therapeutic repertoire is to take advantage of the resident prophages present in many M. abscessus strains. Bioinformatic analysis shows that about 75% of M. abscessus strains contain at least one integrated prophage [43], and a new computational tool (DEPhT) assists in identifying these [44]. They are genomically distinct from the M. smegmatis phages, and are highly diverse, but can be grouped into clusters according to their relatedness (MabA, MabB etc.) [43]. Most are bioinformatically predicted to be intact, and thus capable of undergoing lytic induction and releasing intact viral particles. Such release can occur spontaneously, such that liquid cultures of most M. abscessus strains are spewing phage particles into the growth medium, and this release is likely to be happening in patients as well. For these to be useful it is first necessary to identify a second strain that these particles will productively infect and form plaques on solid medium. Large pairwise screens successfully identified many new phages, which could then be purified, analyzed, and further characterized [36, 43]; none of these infect M. smegmatis, consistent with their genomic dissimilarities [36, 43]. Screening of these against an extant M. abscessus collection showed several instances of strains that are infected by these that are not infected by any of the M. smegmatis phages [36]. The range could be potentially expanded further through the isolation of host range mutants as described above. However, prior to therapeutic use, they need to be engineered to be lytic and to meet other FDA criteria. This is complicated because the engineering and propagation for therapeutic use needs to be done in an M. abscessus strain, presenting logistical and technical challenges. Nonetheless, this may represent the most encouraging direction for achieving broader coverage of the rough M. abscessus strains with phage cocktails.
An important consideration in these approaches is the choice of bacterial strains for propagating phages for therapy. The advantage of using M. smegmatis mc2155 is that it is prophage-free, avoiding contamination of phage preparations with spontaneously induced prophages. The potential advantage of using a specific M. abscessus strain for phage propagation is that it may help to circumvent constraints such as restriction-modification systems. And because none of the lytically growing phages recovered from M. abscessus prophages described above infect M. smegmatis, these need to be prepared using M. abscessus. However, using M. abscessus for large scale preparation is challenging because of biosafety concerns and the need for prophage-free strains sensitive for that phage.
Finally, can phages be identified for killing smooth morphotype M. abscessus strains? It is helpful to first define the root of the problem. Smooth strains have abundant GPLs in their cell walls, and these could block phage attachment or receptor recognition. This may constrain the numbers and types of phages that infect these, but it is not the whole issue. Several smooth strains are infected quite well by some phages as seen by plaque formation, but when challenged by the lytic phages in liquid culture, killing is extremely poor and does not warrant therapeutic use (Fig. 3C). This could result from a post-DNA injection blockage that interferes with killing but not infection per se, although its mechanism and relationship to GPLs is not understood. We note that this phenotype of infection without killing is also observed for a few of the rough strains [36]. Circumventing these two apparent barriers might involve the isolation of additional phages selected directly on smooth strains, construction of recombinant phages that can recognize their receptors without GPL interference or identifying ways to circumvent the post-injection block.
Prospects for Phage Therapy of Mycobacterium Infections
The current use of phages for Mycobacterium therapies is constrained to compassionate use for NTM infections. These have provided considerable insights into safety, immunogenicity, and resistance, as well as potential biomarkers that can be used to monitor patient responses [35, 38]. However, each case is complicated with multiple confounding factors and often concurrent infections. Treating patients earlier after diagnosis would likely help to minimize these complications and provide clarity as to the potential benefits of therapy. Providing phages for personalized single cases is unlikely to be sustainable for the longer term but may provide a bridge to formal clinical trials that can begin to identify critical parameters and learn how the many variables influence outcomes. Developing such trials is non-trivial because of the challenging patient population and the considerable costs involved. Manufacturing of phages using good manufacturing practice (GMP) remains cost-prohibitive in most instances. The recent clinical trials started elsewhere for phage therapies of Pseudomonas in cystic fibrosis patients provides encouragement that NTM trials may not be so far off.
Most of the compassionate use cases for NTM infections have been targeted at M. abscessus infection. However, there are other important Mycobacterium pathogens that are in need of alternative therapies. In the US, Mycobacterium avium complex (MAC) infections are quite prevalent among older women [45] and are typically a challenge to treat with antibiotics [46, 47]. Other NTM pathogens are less common, including Mycobacterium xenopi, Mycobacterium chimera, Mycobacterium chelonae, and Mycobacterium kansasii, but can be difficult to treat as well [48, 49]. Phages have been used to treat a small number of patients with some of these but, like M. abscessus, these strains typically show strong variations in phage susceptibility and require personalization.
M. tuberculosis infections present an interesting contrast to the NTM cases. The disease is very prevalent, although mostly confined to developing countries. It is treatable with multiple drug therapies for an extended period of time (6 months), but non-compliance is common and antibiotic resistance develops relatively rapidly. There is thus a need for approaches that shorten current therapies, enhance compliance, and reduce resistance, for treatment of highly drug resistant strains, and to help protect newly developed (and often very expensive) drugs from resistance [14]. Phages could potentially play roles in these strategies [14].
M. tuberculosis clinical isolates are genetically more homogenous than M. abscessus and other NTM strains, and there is much less variability in the phage infection profiles [23]. A relatively small cocktail of 3–5 phages infects multiple isolates of the major M. tuberculosis lineages, with two or more phages infecting each strain [23]. Thus, personalization can likely be avoided, which is important for a pathogen that takes so long to grow and to test for phage infection profiles. Although there is a broad need for new therapies, compassionate use cases to treat M. tuberculosis infections are rare as there are often newly developed antibiotics that may be early in the approval process but can be considered for treatment. However, clinical trials testing for antimycobacterial activity can be envisaged that might be instructive about the prospects for treatment. Because of the complexities of the disease, with an intracellular bacterial population and often well-developed granulomas, optimism should be tempered in the absence of clinical evidence, as it is not clear whether phages can access intracellular bacteria. However, the possibility of at least reducing the extracellular population of bacteria by phages in combination with antibiotics warrants investigation.
References
Abedon ST. Use of phage therapy to treat long-standing, persistent, or chronic bacterial infections. Adv Drug Deliv Rev. 2019;145:18–39.
Summers WC. Felix d’Herelle and the origins of molecular biology. New Haven: Yale University Press; 1999.
Chanishvili N. Phage therapy–history from Twort and d’Herelle through Soviet experience to current approaches. Adv Virus Res. 2012;83:3–40.
Lusiak-Szelachowska M, Miedzybrodzki R, Drulis-Kawa Z, Cater K, Knezevic P, Winogradow C, et al. Bacteriophages and antibiotic interactions in clinical practice: what we have learned so far. J Biomed Sci. 2022;29(1):23.
Zaldastanishvili E, Leshkasheli L, Dadiani M, Nadareishvili L, Askilashvili L, Kvatadze N, et al. Phage therapy experience at the Eliava Phage Therapy Center: three cases of bacterial persistence. Viruses. 2021;13(10):1901.
Hatfull GF, Dedrick RM, Schooley RT. Phage therapy for antibiotic-resistant bacterial infections. Annu Rev Med. 2022;73:197–211.
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55.
Degiacomi G, Sammartino JC, Chiarelli LR, Riabova O, Makarov V, Pasca MR. Mycobacterium abscessus, an emerging and worrisome pathogen among cystic fibrosis patients. Int J Mol Sci. 2019;20(23):5868.
van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat. 2012;15(3):149–61.
Schooley RT, Strathdee S. Treat phage like living antibiotics. Nat Microbiol. 2020;5(3):391–2.
To K, Cao R, Yegiazaryan A, Owens J, Venketaraman V. General overview of nontuberculous mycobacteria opportunistic pathogens: Mycobacterium avium and Mycobacterium abscessus. J Clin Med. 2020;9(8):2541.
Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. 2012;67(4):810–8.
Shield CG, Swift BMC, McHugh TD, Dedrick RM, Hatfull GF, Satta G. Application of bacteriophages for mycobacterial infections, from diagnosis to treatment. Microorganisms. 2021;9(11):2366.
Diacon AH, Guerrero-Bustamante CA, Rosenkranz B, Rubio Pomar FJ, Vanker N, Hatfull GF. Mycobacteriophages to treat tuberculosis: dream or delusion? Respiration. 2022;101(1):1–15.
Hatfull GF. Mycobacteriophages. Microbiol Spectr. 2018. https://doi.org/10.1128/microbiolspec.GPP3-0026-2018.
Hatfull GF. Actinobacteriophages: genomics, dynamics, and applications. Annu Rev Virol. 2020;7(1):37–61.
Hatfull GF. Mycobacteriophages: from petri dish to patient. PLoS Pathog. 2022;18(7): e1010602.
Hashemi Shahraki A, Mirsaeidi M. Phage therapy for Mycobacterium abscessus and strategies to improve outcomes. Microorganisms. 2021;9(3):596.
Senhaji-Kacha A, Esteban J, Garcia-Quintanilla M. Considerations for phage therapy against Mycobacterium abscessus. Front Microbiol. 2020;11: 609017.
Sullivan MR, Rubin EJ, Dulberger CL. Mycobacteriophages as genomic engineers and anti-infective weapons. MBio. 2021. https://doi.org/10.1128/mBio.00632-21.
Dedrick R, Guerrero Bustamante C, Garlena RA, Russell DA, Ford K, Harris K, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25:730–3.
Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, et al. On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434(2):187–201.
Guerrero-Bustamante CA, Dedrick RM, Garlena RA, Russell DA, Hatfull GF. Toward a phage cocktail for tuberculosis: susceptibility and tuberculocidal action of mycobacteriophages against diverse Mycobacterium tuberculosis strains. MBio. 2021. https://doi.org/10.1128/mBio.00973-21.
Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, et al. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology. 2009;155(Pt 9):2962–77.
Dedrick RM, Guerrero Bustamante CA, Garlena RA, Pinches RS, Cornely K, Hatfull GF. Mycobacteriophage ZoeJ: a broad host-range close relative of mycobacteriophage TM4. Tuberculosis (Edinb). 2019;115:14–23.
Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3(12): e3957.
Wetzel KS, Guerrero-Bustamante CA, Dedrick RM, Ko CC, Freeman KG, Aull HG, et al. CRISPY-BRED and CRISPY-BRIP: efficient bacteriophage engineering. Sci Rep. 2021;11(1):6796.
Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7(3):395–405.
Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. Inquiry learning. Teaching scientific inquiry. Science. 2006;314(5807):1880–1.
Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio. 2014;5(1):e01051-e1113.
Hanauer DI, Graham MJ, Sea P, Betancur L, Bobrownicki A, Cresawn SG, et al. An inclusive Research Education Community (iREC): impact of the SEA-PHAGES program on research outcomes and student learning. Proc Natl Acad Sci USA. 2017;114(51):13531–6.
Russell DA, Hatfull GF. PhagesDB: the actinobacteriophage database. Bioinformatics. 2017;33(5):784–6.
Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife. 2015;4: e06416.
Pope WH, Montgomery MT, Bonilla JA, Dejong R, Garlena RA, Guerrero Bustamante C, et al. Complete genome sequences of 38 Gordonia sp. bacteriophages. Genome Announc. 2017. https://doi.org/10.1128/genomeA.01143-16.
Dedrick RM, Smith BE, Cristinziano M, Freeman KG, Jacobs-Sera D, Belessis Y, et al. Phage therapy of Mycobacterium infections: compassionate-use of phages in twenty patients with drug-resistant mycobacterial disease. Clin Infect Dis. 2022. https://doi.org/10.1093/cid/ciac453.
Dedrick RM, Smith BE, Garlena RA, Russell DA, Aull HG, Mahalingam V, et al. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. MBio. 2021. https://doi.org/10.1128/mBio.03431-20.
Gutierrez AV, Viljoen A, Ghigo E, Herrmann JL, Kremer L. Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front Microbiol. 2018;9:1145.
Nick JA, Dedrick RM, Gray AL, Vladar EK, Smith BE, Freeman KG, et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell. 2022;185(11):1860–74.
Little JS, Dedrick RM, Freeman KG, Cristinziano M, Smith BE, Benson CA, et al. Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nat Commun. 2022;13(1):2313.
Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.00954-17.
van Kessel JC, Hatfull GF. Mycobacterial recombineering. Methods Mol Biol. 2008;435:203–15.
Marinelli LJ, Hatfull GF, Piuri M. Recombineering: a powerful tool for modification of bacteriophage genomes. Bacteriophage. 2012;2(1):5–14.
Dedrick RM, Aull HG, Jacobs-Sera D, Garlena RA, Russell DA, Smith BE, et al. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. MBio. 2021. https://doi.org/10.1128/mBio.03441-20.
Gauthier CH, Abad L, Venbakkam AK, Malnak J, Russell DA, Hatfull GF. DEPhT: a novel approach for efficient prophage discovery and precise extraction. Nucleic Acids Res. 2022;50(13): e75.
Jamal F, Hammer MM. Nontuberculous mycobacterial infections. Radiol Clin North Am. 2022;60(3):399–408.
Kadota T, Matsui H, Hirose T, Suzuki J, Saito M, Akaba T, et al. Analysis of drug treatment outcome in clarithromycin-resistant Mycobacterium avium complex lung disease. BMC Infect Dis. 2016;16:31.
Moon SM, Park HY, Kim SY, Jhun BW, Lee H, Jeon K, et al. Clinical characteristics, treatment outcomes, and resistance mutations associated with macrolide-resistant Mycobacterium avium complex lung disease. Antimicrob Agents Chemother. 2016;60(11):6758–65.
Chen LC, Huang HN, Yu CJ, Chien JY, Hsueh PR. Clinical features and treatment outcomes of Mycobacterium chimaera lung disease and antimicrobial susceptibility of the mycobacterial isolates. J Infect. 2020;80(4):437–43.
Zaheen A, Hirama T, Mehrabi M, Brode SK, Marras TK. Clinical outcomes in Mycobacterium xenopi versus Mycobacterium avium complex pulmonary disease: a retrospective matched cohort study. Respir Med. 2020;167: 105967.
Acknowledgements
Funding
This work was supported by National Institutes of Health grant GM131729 and Howard Hughes Medical Institute Grant GT12053.
Editorial Assistance
I would like to especially thank the numerous colleagues we have worked with in providing compassionate use therapies, and the bravery of the patients who have agreed to these experimental treatments. Drs Benson and Schooley at IPATH have provided critical guidance in these treatments which would not have happened without them. Students and faculty in the SEA-PHAGES program have fueled phage discovery and advanced these therapies. I thank members of my laboratory for their marvelous contributions, including Owen Abad, Haley Aull, Maddie Cristinziano, Bekah Dedrick, Krista Freeman, Becky Garlena, Christian Gauthier, Carlos Guerrero, Debbie Jacobs-Sera, Ari Kaganovsky, Ching-Ching Ko, Crystal Petrone, Dan Russell, Bailey Smith, and Katie Wetzel. I am grateful to Owen Abad, Maddie Cristinziano, Krista Freeman, and Carlos Guerrero for critical comments on the manuscript.
Author Contributions
Graham F. Hatfull wrote the article.
Disclosures
Graham F. Hatfull has a Collaborative Research Agreement with Janssen Inc.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by the author.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Hatfull, G.F. Phage Therapy for Nontuberculous Mycobacteria: Challenges and Opportunities. Pulm Ther 9, 91–107 (2023). https://doi.org/10.1007/s41030-022-00210-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41030-022-00210-y