Abstract
Machine learning (ML) is a discipline of computer science in which statistical methods are applied to data in order to classify, predict, or optimize, based on previously observed data. Pulmonary and critical care medicine have seen a surge in the application of this methodology, potentially delivering improvements in our ability to diagnose, treat, and better understand a multitude of disease states. Here we review the literature and provide a detailed overview of the recent advances in ML as applied to these areas of medicine. In addition, we discuss both the significant benefits of this work as well as the challenges in the implementation and acceptance of this non-traditional methodology for clinical purposes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Machine learning is a sub-discipline of computer science that is becoming widely utilized across the medical field. |
This methodology is being applied to many areas of pulmonary and critical care medicine, and these fields potentially have much more to gain from the use of machine learning. |
Chest imaging analysis, pulmonary function test interpretation, and sepsis analytics are some examples of topics in which these methods have the potential to lead to significant diagnostic and therapeutic improvements. |
It is important that all clinicians and researchers now begin to understand both the potential benefits as well as the challenges and limitations of machine learning in medical research. |
Introduction
In recent years, there have been great advances in the use of machine learning (ML) in medicine. Through the use of large clinical databases, researchers have tackled previously unanswerable questions, and created systems that augment human decision-making skills [1, 2]. This exciting area of research has historically experienced cycles in which enthusiasm and optimism alternate with skepticism and pessimism. While there is good reason to be wary of some of the grandiose claims currently being made regarding this kind of work [3], ML-based models have already proven to be effective tools for selected clinical purposes [4]. The research required to expand these methods across different areas and uses in medicine is currently in a dramatic growth stage; while we cannot be certain what role ML will ultimately play in healthcare, it is very likely to become progressively more fully ingrained into the future practice of medicine. It is therefore important that clinicians and researchers understand the basics of ML, and how it can, cannot, and should not be used for both research and applied clinical purposes.
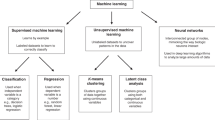
ML is a sub-discipline of computer science in which computers ‘learn’ from large quantities of data in order to find patterns without being explicitly programmed to do so [1, 2]. One way it differs from traditional statistical analysis is that the main goal of ML is accurate prediction, classification, or optimization, so that understanding the associations between the variables is not as relevant, and generally not as transparent, as with more complex algorithms [5]. There are two main types of ML—supervised and unsupervised learning. Supervised learning occurs when inputs are chosen according to some pre-defined output: the development of predictive models that can identify suspicious pulmonary nodules on chest imaging provides an example [6]. Unsupervised learning finds patterns among data without using a pre-specified label [5]. This method has been employed to identify different phenotypes of sepsis [7]. There are many different subclassifications of these two broad concepts, the details of which are beyond the scope of this review.
ML has already made its way into the domain of pulmonary and critical care medicine. It is being applied to imaging technology to enhance the way we screen and manage pulmonary nodules [6], as well as aiding in pulmonary function test (PFT) analysis, and the diagnosis of, and exacerbation prediction in chronic obstructive pulmonary disease (COPD) [8]. Researchers using ML have pioneered the development of illness severity scores in the intensive care unit (ICU) [9, 10]. More recent studies have leveraged the abundance of data present in the ICU to predict the course of mechanical ventilation and the occurrence of sepsis, as well as to support decision making in this context (e.g., fluid vs. vasopressor choices in sepsis care) [11,12,13,14,15,16,17]. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Methods of Literature Selection
The literature search was conducted in (PubMed). Research papers, systematic reviews, and narrative reviews published prior to November 1, 2019 were included. Non-English papers, abstracts without full text, and pediatric studies were excluded. The search terms used to find relevant literature included: (“machine learning” OR “deep learning” OR “neural network” OR “artificial intelligence”) AND (“intensive care unit” OR “ICU” OR “critical care” OR “pulmonary” OR “lung” OR “COPD” OR “sepsis”).
A total of 2502 papers were initially identified with these search terms, of which 108 pediatric studies were excluded, leading to a final count of 2394. More focused searches of pulmonary specific literature and critical care-specific literature revealed 694 papers and 1835 papers, respectively. There were 135 papers duplicated between these two categories. Two systematic reviews were identified [18, 19]. Given the narrative nature of this review, the final cohort of papers was hand-picked to provide the reader with the best general overview of the topic and was not meant to be comprehensive. We selected 19 research manuscripts and one systematic review (Table 1), and referenced a number of narrative reviews.
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Analyzing Chest Imaging
One of the most successful clinical applications of ML to date is that of imaging analysis. As of January 2019, there were 14 FDA-approved medical artificial intelligence (AI) technologies, most involving imaging. These included a software application for the detection of cerebral hemorrhage on computed tomography (CT) scan, a retinal scanner that can detect diabetic retinopathy, and a mammography tool that works to classify breast density [4]. In the world of pulmonary medicine, chest imaging plays an integral role in diagnosis and long-term management. Chest radiographs and chest CTs have been intensely studied, with start-up companies now offering automated image analysis [20].
Much of the ML for image analysis, including that of chest radiographs and CTs, involves a type of deep learning that employs convolutional neural networks (CNN). The use of the word ‘deep’ in this context does not imply profundity, but simply describes the depth in layers (i.e., multiple) of the network being trained. All images contain millions of pixels which need to be processed by these systems in order to be able to recognize patterns, and CNNs make this process more efficient by segmenting an image and avoiding the need to process each pixel separately. Like the hierarchical structure of the visual system in the human brain, the network of CNNs is layered in units that detect specific features. The output of these units is iteratively fine-tuned (changing the ‘weights’ assigned to particular inputs) to reduce the error as closely as possible based on the data used for training [21, 22].
One of the earliest studies that applied CNNs to radiographs was that of Lakhani et al. who used two different CNN models on four separate datasets to develop a model that could diagnose pulmonary tuberculosis. Remarkably, the best performing model had an area under the receiver operating curve (AUC) of 0.99 [23]. In 2018, Rajpurkar et al. developed a CNN called CheXNeXt, which was developed to recognize 14 different pulmonary pathologies on chest radiographs. The algorithm performed equally well as radiologists in diagnosing ten pathologies, better in one pathology (atelectasis), and worse in three pathologies (cardiomegaly, emphysema, and hiatal hernia) [24]. There is particular potential for the use of these models in resource-poor settings where a radiologist may not be available.
Another area that has received great interest is the detection and classification of pulmonary nodules via CT scan. There are algorithms that assist with the detection, per se, of nodules (computer-aided detection or CAD), and those that assist with specific diagnosis (computer-aided diagnosis or CADx) [25]. The extent of the work in CAD is reflected in the 2019 systematic review by Pehrson et al., which presented 41 papers on ML models for automated nodule detection applied to the Lung Image Database Consortium Image Collection. These studies are heterogenous in their approach to modeling, with some using CNN or other “deep learning” methods, while others use more “feature-based” approaches. The feature-based approaches require that the images already have structured annotations to describe the nodules, whereas the deep learning algorithms do not require this step, making the latter more generalizable. In the Pehrson review, five of the feature-based and two of the deep learning methods had an accuracy of more than 95% for nodule detection [18].
There has also been extensive research into CADx, where the usual goal is to develop models that can distinguish between benign and malignant lesions based on imaging. Uthoff et al. captured specific perinodular parenchymal features in a model which showed significant improvement compared to a model using only nodular features. This was the first study to report a comparison to the Fleischner pulmonary nodule follow-up guidelines, and suggested a reduction in repeat imaging and biopsy is possible with the use of the algorithm [6]. A 2019 paper presented a deep learning algorithm that employed prior and current CT scan lung volumes to predict the risk of malignancy. When previous images were available to view, the algorithm performed similarly to radiologists, but actually outperformed the radiologists when initial images were not available [26].
ML has also been applied to histopathology to help improve the accuracy and efficiency of diagnosis and prognostication of non-small cell lung cancer (NSCLC) [25]. Koh et al. developed a three-marker immunohistochemistry panel (TTF‐1, Napsin A, and p40) that could be used with minimal tissue samples of ambiguous morphology, and succeeded in differentiating adenocarcinoma from squamous cell carcinoma in 82% of cases. In those cases where the initial panel was non-diagnostic, the use of two additional markers (p63 and CK5/6) provided the ability to further subtype 72% of the cases [27]. This is just one of the many examples of the work in this field, and as more high-throughput technologies are developed for slide analysis, the expectation is that clinicians will be able to get more accurate data more efficiently with less tissue required [23].
Chronic Obstructive Pulmonary Disease and Pulmonary Function Tests
COPD is a leading cause of morbidity and mortality worldwide, and a significant burden on both patient quality of life and the healthcare system [28]. ML is being used to help enhance the management of this disease through improved PFT analysis as well as prediction of COPD exacerbations [8].
PFT analysis is an area of medicine where AI may be extraordinarily beneficial to clinicians. In one study, the authors tested their algorithm against 16 pulmonologists in 50 cases, using the American Thoracic Society/European Respiratory Society (ATS/ERS) guidelines as the gold standard. The pulmonologists interpreted the PFTs accurately in 75% of cases and established the correct diagnosis in only 44% of the cases with a large inter-rater variability (kappa 0.35). In contrast, the computer algorithm correctly interpreted 100% of the PFTs and established the correct diagnosis in 82% of the cases [29]. Another study looked at nearly 1000 patients in an outpatient practice who were diagnosed with different pulmonary diseases (including COPD) based on PFTs and other studies performed at a clinician’s discretion. A decision tree model was developed that combined PFT results with patient characteristics, and was compared against a modified ATS/ERS algorithm for PFT interpretation. The model had a 68% accuracy (vs. 38% for the ATS/ERS algorithm) for choosing the correct diagnosis; it also displayed significant improvements in positive predictive value and sensitivity in the detection of four disease states, including COPD [30]. This demonstrates that ML has the potential to improve the automation of PFT interpretation, although it is not currently a replacement for human input and decision making. The accuracy of interpretations may be further enhanced by incorporating contextual patient-specific features [8].
There has also been a great deal of work done in the prediction and prevention of COPD exacerbations. Remote monitoring of high-risk patients with COPD has been employed to reduce the number of COPD exacerbations requiring hospitalization, but has largely been unsuccessful with many false-positive cases reported. A 2018 study looked at how applying ML to remote monitoring data could help improve prediction accuracy for this particular purpose. These researchers employed data collected for the Telescot COPD trial, with the ML algorithm outperforming previously used models in both prediction of COPD exacerbation and the need for corticosteroids [31].
Another study employed physiologic data along with ambulatory recording of respiratory sounds to predict COPD exacerbations. The pilot study of 15 patients allowed development of a model that could predict symptom-based episodes 4.4 days prior to an exacerbation. The accuracy of the model was 78% (32 of 41 exacerbations detected), with only a 2.2% false-positive rate. This study has its limitations in terms of the small sample size and the use of only one symptom-based criterion, but this relatively simple approach demonstrates the potential for more robust work in this area [32].
Critical Care Medicine
Tests and treatments are difficult to evaluate among critically ill patients because of marked disease heterogeneity in the ICU. The vast majority of randomized controlled trials (RCTs) for new therapies in this population have yielded negative results. This is at least in part due to the complexity of the disease processes in question, as well as significant population heterogeneity, inadequate sample sizes, and overly optimistic expected effects [33]. ML approaches have been applied to study this patient population in a manner that can overcome some of the limitations of traditional RCTs. For example, ML can supply the prediction of patient subsets who are more likely to benefit from (or who are more likely to be harmed by) a test or treatment—the holy grail not just of critical care medicine but all of healthcare [34].
A recent development that has enabled and propelled the application of ML to medicine is the adoption of electronic health records (EHR). In the United States, the Health Information Technology for Economic and Clinical Health (HITECH) Act has allowed for the rapid implementation of EHR nationwide, and this has had a global impact [35]. In the field of intensive care, a National Institutes of Health-funded initiative entitled Medical Information Mart for Intensive Care (MIMIC), developed by a partnership between Beth Israel Deaconess Medical Center (BIDMC), Massachusetts Institute of Technology (MIT) and Philips Healthcare, has led to rapid advancement in the application of data science to clinical medicine [36]. MIMIC is a publicly available, de-identified database of close to 100,000 patients admitted to the ICUs at BIDMC in Boston, MA, USA from 2002 to 2018 and is maintained by the Laboratory for Computational Physiology at MIT [37]. Access has been granted to more than 12,000 registered users who provide proof of training in human subject research and adhere to a data user agreement. In addition to MIMIC, the MIT group also maintains a public repository of reusable codes and queries to extract common ICU concepts in order to promote reproducibility and to allow investigators to build on each other’s research [38]. MIMIC and other datasets have allowed clinicians, researchers, and data scientists from across the world to explore the use of ML for classification, prediction, and optimization to help improve the management of critically ill patients [34].
Sepsis
Sepsis is one condition that has been a major focus of both basic and clinical research for many years in an attempt to understand the underlying pathophysiology and risk factors, as well as to identify the best methods of treatment. ML is being implemented from multiple fronts in order to tackle some of these fundamental questions about sepsis [13,14,15, 34].
A very early study in this field did not make use of a large dataset but rather studied a select sample of 92 patients. This 2008 work employed seven cytokine biomarkers and a neural network approach to develop a predictive model for sepsis with a positive predictive value of 92% and a negative predictive value of 80%. The model outperformed other models based on clinical parameters alone [13]. Wang et al. extracted clinical EHR data and employed a support vector machine to predict which patients will develop severe sepsis or septic shock with a high sensitivity and specificity [14]. Nemati et al. developed the Artificial Intelligence Sepsis Expert (AISE), which incorporates EHR data with high-resolution blood pressure and heart rate measurements to predict the onset of sepsis up to 12 h prior to meeting diagnostic criteria. The AUC of this model was 0.85 at 4 h prior and 0.83 at 12 h prior to onset time [15].
The treatment of sepsis and septic shock are also targets for improvement through a more individualized approach that employs ML, particularly in decisions regarding the administration of fluids versus vasopressors to correct hemodynamic instability. The use of a reinforcement learning model in a study by Komorowski et al. sought to do just that. In this study, the model was trained on over 17,000 unique cases and was optimized for improving mortality. It was found that mortality was lowest when the actual clinical decision matched the model’s suggestion, and that mortality increased with fluid or vasopressor in excess of the suggested dose [16]. An additional study in 2019 sought to predict changes in urine output of septic patients in response to fluid administration, an important surrogate of end-organ perfusion. This model, based on a gradient boosting algorithm, had an AUC of 0.86 for predicting decreased urine output after fluid administration, and was most sensitive (92.2%) at predicting persistent oliguria in patients with oliguria preceding fluid administration [17]. It is clear from this work that there is great potential in ML methods to improve how we manage sepsis and predict volume responsiveness in the ICU.
ML is also being used to help gain a better understanding of the complexity and heterogeneity of many pertinent conditions through the discovery of different clinical or biochemical phenotypes. Sepsis is a heterogenous disease, yet current guidelines dictate a similar therapeutic approach across all patients. The same is true with many other diseases in the ICU including acute respiratory distress syndrome [39]. In order to provide more precise therapy, the nature of this heterogeneity must be better understood.
A 2019 study sought to better define these differences among patients with sepsis. The authors analyzed data from over 60,000 patients with sepsis who were previously enrolled in large RCTs. Using unsupervised learning, they discovered four distinct clinical phenotypes, each correlating with different patient-specific features, biomarker patterns, and clinical outcomes. They then ran simulations of the RCTs and found marked differences in the effect size of the interventions depending on the phenotypic make-up of the patients enrolled [7]. The results of this study will undoubtedly drive more work to further characterize increasingly precise phenotypes in order to inform the design of RCTs and potentially suggest targeted individualized treatments.
Mechanical Ventilation
Mechanical ventilation is another area in critical care that has been the subject of ML models. A 2018 study using MIMIC looked at 20,000 patients who were mechanically ventilated in the ICU, and developed a model to predict which patients would require prolonged mechanical ventilation (PMV) and a tracheostomy procedure. The AUCs of the model for PMV and tracheostomy were 0.82 and 0.83, respectively. They found that the most relevant features were the pulmonary logistic organ dysfunction scores (LODS) for PMV, and, unexpectedly, a diagnosis of cardiac arrhythmia for tracheostomy. These models may have important implications for prognostication, and have the potential to improve outcomes by proceeding to tracheostomy sooner in select patients, thereby facilitating earlier mobilization [11].
Another aspect of mechanical ventilation is readiness for extubation. Hsieh et al. studied over 3000 patients and employed a neural network approach to predict which extubated patients would require re-intubation within 72 h. The model had an AUC of 0.85, which compared favorably to the current gold standard, the rapid shallow breathing index, which registered an unimpressive AUC of 0.54 in the same cohort [12].
Severity of Illness Scoring Systems
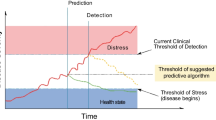
The final topic to be addressed is illness severity scoring. These scores, such as the Acute Physiology and Chronic Health Evaluation (APACHE) and the Simplified Acute Physiology Score (SAPS), are often used as quality metrics for hospital outcomes and for observational research, but they have not proven to be nearly as useful at the front lines of clinical practice. These scores do not reflect the dynamic and heterogenous nature of the critically ill patient, particularly those at highest risk of mortality. Additionally, data from the first day of admission are utilized in these scores so that the subsequent course of events is not taken into consideration. It also would be valuable to clinicians to have an earlier, more immediate assessment of risk of mortality. A more clinically applicable score could have major implications for patients and their families with regards to more accurate prognostication and realistic goals of care conversations [10].
Awar et al. utilized multiple ML methods on the MIMIC dataset to create a more reliable scoring system available at 6-h post-admission. Their ensemble learning random forest model showed the most robust performance profile, outperforming all standard scoring systems by way of AUC [9]. Cosgiff et al. similarly leveraged a variety of ML methods to develop a system calibrated to predict outcomes reliably across all risk groups, with the hypothesis that a sequential modeling approach would help achieve this. They compared their methods to the APACHE IV system. They were able to show some improvement in AUC for their sequential modeling and gradient boosting model, but the effect size was only modest [10]. While these ML-based algorithms do improve upon the current standard, there is still more work to be done to improve predictive capacity and generalizability.
Discussion: Challenges and Next Steps
There are several limitations to these studies and more work is required to safely implement these models into actual clinical decision making. First, many of these studies developed and validated the models within the same patient cohort; therefore, generalizability to other populations has not been evaluated. Second, the models have yet to be tested prospectively and their benefit on clinical or other outcomes demonstrated. Third, healthcare organizations currently lack the expert human resources and the information technology or IT capabilities to implement—evaluate, incorporate, and continuously monitor and re-calibrate—ML into clinical practice [3]. Fourth, there remains significant trust issues around the collection and safe use of digital data that need to be addressed [40]. Fifth, at this moment, most healthcare systems worldwide are likely just not sufficiently robust to easily absorb these technologies into current workflows. The healthcare systems that we know today were not designed for a world of augmented decision making and predictive modeling, and will require a number of enhancements to successfully incorporate these kinds of innovative advances.
From a policy perspective, there are uncertainties concerning where, when, and which regulations affecting ML will be required. There are also no current definitions of acceptable performance standards, accuracy rates, and acceptable patient outcomes against which to measure the algorithms. Most importantly, however, the impact of ML will be negated if hospitals and clinics simply lack the capacity to effectively treat high-risk patients or implement population health interventions due to technical and workflow limitations.
There is reason to be optimistic, however. In March 2019, the World Health Organization established a digital health department whose aim is to harness “the power of digital health and innovation to assess, integrate, regulate, and maximize the opportunities of digital technologies and artificial intelligence” [41]. Across the globe, governments are partnering with universities and industry to build an ML roadmap. Programs and events bring together clinicians, computer scientists, and engineers to create a collaborative ecosystem that can leverage the power of data science [42]. These cross-disciplinary partnerships are crucial in order to design ML in a manner that is usable, safe, and trustworthy for all the humans touched by its applications [43].
Conclusions
ML and the broader notion of AI have already proven to be valuable in the medical field, and have the potential to lead to sweeping changes in how we practice medicine. In pulmonary and critical care medicine, there has been great effort in applying these methods in order to improve upon our ability to diagnose and treat patients accurately and efficiently, with the overall goal of improving outcomes for our patients. The specific areas in this field which have seen the most utility in applying these methods are in pulmonary imaging and nodule detection, COPD and PFT interpretation, and predictive modeling for critically ill patients. The scope of research is likely to progress in the coming years to become inclusive of many more facets of pulmonary and critical care. The use of ML in this context is not without its challenges and pitfalls, and we must focus on addressing these problems in order to ensure the safety of our patients, promote only the highest-quality research, and help foster a more universal acceptance and implementation of this methodology. In doing so, we will be able to see the true potential of ML in medicine come to fruition.
References
Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–30.
Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319(13):1317–8.
Panch T, Mattie H, Celi LA. The “inconvenient truth” about AI in healthcare. Npj Digital Medicine. 2019;2(1):1–3.
Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56.
Sidey-Gibbons JA, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19(1):64.
Uthoff J, Stephens MJ, Newell Jr JD, Hoffman EA, Larson J, Koehn N, De Stefano FA, Lusk CM, Wenzlaff AS, Watza D, Neslund–Dudas C. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Medical Physics. 2019.
Seymour CW, Kennedy JN, Wang S, Chang CC, Elliott CF, Xu Z, Berry S, Clermont G, Cooper G, Gomez H, Huang DT. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321(20):2003–17.
Franssen FM, Alter P, Bar N, Benedikter BJ, Iurato S, Maier D, Maxheim M, Roessler FK, Spruit MA, Vogelmeier CF, Wouters EF. Personalized medicine for patients with COPD: where are we? Int J Chronic Obstr Pulm Dis. 2019;14:1465.
Awad A, Bader-El-Den M, McNicholas J, Briggs J. Early hospital mortality prediction of intensive care unit patients using an ensemble learning approach. Int J Med Informatics. 2017;108:185–95.
Cosgriff CV, Celi LA, Ko S, Sundaresan T, de la Hoz MÁ, Kaufman AR, Stone DJ, Badawi O, Deliberato RO. Developing well-calibrated illness severity scores for decision support in the critically ill. NPJ Digital Medicine. 2019;2(1):1–8.
Parreco J, Hidalgo A, Parks JJ, Kozol R, Rattan R. Using artificial intelligence to predict prolonged mechanical ventilation and tracheostomy placement. J Surg Res. 2018;228:179–87.
Hsieh MH, Hsieh MJ, Chen CM, Hsieh CC, Chao CM, Lai CC. An artificial neural network model for predicting successful extubation in intensive care units. J Clin Med. 2018;7(9):240.
Lukaszewski RA, Yates AM, Jackson MC, Swingler K, Scherer JM, Simpson AJ, Sadler P, McQuillan P, Titball RW, Brooks TJ, Pearce MJ. Presymptomatic prediction of sepsis in intensive care unit patients. Clin Vaccine Immunol. 2008;15(7):1089–94.
Wang SL, Wu F, Wang BH. Prediction of severe sepsis using SVM model. In: Advances in computational biology 2010. New York: Springer. pp. 75–81.
Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46(4):547–53.
Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018;24(11):1716.
Lin PC, Huang HC, Komorowski M, Lin WK, Chang CM, Chen KT, Li YC, Lin MC. A machine learning approach for predicting urine output after fluid administration. Comput Methods Programs Biomed. 2019;177:155–9.
Pehrson LM, Nielsen MB, Ammitzbøl Lauridsen C. Automatic pulmonary nodule detection applying deep learning or machine learning algorithms to the LIDC-IDRI database: a systematic review. Diagnostics. 2019;9(1):29.
Linnen DT, Escobar GJ, Hu X, Scruth E, Liu V, Stephens C. Statistical modeling and aggregate-weighted scoring systems in prediction of mortality and ICU transfer: a systematic review. J Hosp Med. 2019;14(3):161.
Lung AI [Internet]. Arterys. [cited 2019Nov1]. Available from: https://www.arterys.com/lung-ai/.
Cosgriff CV, Celi LA. Deep learning for risk assessment: all about automatic feature extraction. Br J Anaesth. 2019.
Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst. 2018;42(11):226.
Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–82.
Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz CP, Patel BN. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15(11):e1002686.
Rabbani M, Kanevsky J, Kafi K, Chandelier F, Giles FJ. Role of artificial intelligence in the care of patients with nonsmall cell lung cancer. Eur J Clin Invest. 2018;48(4):e12901.
Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, Naidich DP. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25(6):954.
Koh J, Go H, Kim MY, Jeon YK, Chung JH, Chung DH. A comprehensive immunohistochemistry algorithm for the histological subtyping of small biopsies obtained from non-small cell lung cancers. Histopathology. 2014;65(6):868–78.
Riley CM, Sciurba FC. Diagnosis and outpatient management of chronic obstructive pulmonary disease: a review. JAMA. 2019;321(8):786–97.
Topalovic M, Das N, Burgel PR, Daenen M, Derom E, Haenebalcke C, Janssen R, Kerstjens HA, Liistro G, Louis R, Ninane V. Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests. Eur Respir J. 2019;53(4):1801660.
Topalovic M, Laval S, Aerts JM, Troosters T, Decramer M, Janssens W, Belgian Pulmonary Function Study investigators. Automated interpretation of pulmonary function tests in adults with respiratory complaints. Respiration. 2017;93(3):170–8.
Orchard P, Agakova A, Pinnock H, Burton CD, Sarran C, Agakov F, McKinstry B. Improving prediction of risk of hospital admission in chronic obstructive pulmonary disease: application of machine learning to telemonitoring data. J Med Internet Res. 2018;20(9):e263.
Fernandez-Granero M, Sanchez-Morillo D, Leon-Jimenez A. Computerised analysis of telemonitored respiratory sounds for predicting acute exacerbations of COPD. Sensors. 2015;15(10):26978–96.
Harhay MO, Wagner J, Ratcliffe SJ, Bronheim RS, Gopal A, Green S, Cooney E, Mikkelsen ME, Kerlin MP, Small DS, Halpern SD. Outcomes and statistical power in adult critical care randomized trials. Am J Respir Crit Care Med. 2014;189(12):1469–78.
Lovejoy CA, Buch V, Maruthappu M. Artificial intelligence in the intensive care unit. BMC Crit Care. 2019;23(1).
Blumenthal D. Implementation of the federal health information technology initiative. N Engl J Med. 2011;365(25):2426–31.
Cosgriff CV, Celi LA, Stone DJ. Critical care, critical data. Biomed Eng Comput Biol. 2019;10:1179597219856564.
Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035.
Johnson AE, Stone DJ, Celi LA, Pollard TJ. The MIMIC code repository: enabling reproducibility in critical care research. J Am Med Inform Assoc. 2017;25(1):32–9.
Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Curr Opin Crit Care. 2019;25(1):12–20.
Copeland R. WSJ News Exclusive | Google’s ‘Project Nightingale’ Gathers Personal Health Data on Millions of Americans [Internet]. The Wall Street Journal. Dow Jones & Company; 2019 [cited 2019 Dec 1]. https://www.wsj.com/articles/google-s-secret-project-nightingale-gathers-personal-health-data-on-millions-of-americans-11573496790.
WHO unveils sweeping reforms in drive towards “triple billion” targets [Internet]. World Health Organization. World Health Organization; [cited 2019 Dec 1]. Available from: https://www.who.int/news-room/detail/06-03-2019-who-unveils-sweeping-reforms-in-drive-towards-triple-billion-targets.
Martineau K. Democratizing artificial intelligence in health care [Internet]. MIT News. 2019 [cited 2019Dec1]. Available from: http://news.mit.edu/2019/democratizing-artificial-intelligence-in-health-care-0118.
Celi LA, Fine B, Stone DJ. An awakening in medicine: the partnership of humanity and intelligent machines. The Lancet Digital Health. 2019;1(6):e255–7.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Eric Mlodzinski and David J. Stone have nothing to disclose. Leo A. Celi is funded by the National Institute of Health through the NIBIB grant R01 EV017205.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced Digital Features
To view enhanced digital features for this article go to https://doi.org/10.6084/m9.figshare.11674023.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any non-commercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mlodzinski, E., Stone, D.J. & Celi, L.A. Machine Learning for Pulmonary and Critical Care Medicine: A Narrative Review. Pulm Ther 6, 67–77 (2020). https://doi.org/10.1007/s41030-020-00110-z
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41030-020-00110-z