Abstract
Patient similarity search is an essential task in healthcare. Recent studies adopted electronic health records (EHRs) to learn patient representations for measuring the clinical similarities. These methods outperformed traditional methods, by capturing more information from various sources consisting of multi-modal EHRs, external knowledge and correlations among medical concepts. They often concerned certain type of data without taking full advantage of various information. We propose a graph representation learning framework, denoted by One-Size-Fits-Three (OSFT), that takes into account fusion-attention, neighbor-attention and global-attention from three types of information. Extensive experiments are conducted on two real datasets of MIMIC-III and MIMIC-IV, and the results verified the effectiveness and generality of our framework. When compared with baselines on patient similarity search, our framework achieved good effectiveness and comparative efficiency. The results provide new insights about whether the use of various information can better measure the patient similarity. The source codes are available at https://github.com/emmali808/ADDS/tree/master/EHRDeepHelper.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The heterogeneous EHRs provide valuable information for evaluating patient similarity, which enables deep learning to show impressive results in downstream applications [31, 40], such as assisted diagnosis and treatment recommendation. Due to the heterogeneous and sparse nature of EHRs, patient similarity learning is still a challenging task. Existing work can be categorized into three groups by using various data: multi-modal EHRs, external knowledge and correlations among medical concepts. The first group adopted EHRs to learn the patient representations [14, 36]. Early work [14] considered textual similarities between EHRs, but showed limited performance on downstream tasks. Later work [36] outperformed early work by using multi-modal information for representation learning. However, due to the sparse nature of EHRs, these methods suffered from the data insufficiency problem by capturing limited content information [27]. To capture more information, the second group introduced external sources, such as medical ontology [6, 27, 34] and medical concept descriptions [26]. They incorporated external knowledge into learning the medical concept embeddings. However, the external sources were limited in terms of quantity and quality [8]. The third group captured hidden medical information such as correlations among medical concepts. MiME [8] modeled the EHRs as hierarchical graphs and employed neural networks to learn the patient representations. However, the EHRs do not always contain complete structure information [9]. Thus, GCT [9] jointly learned the hidden structure while performing various prediction tasks. However, these models were reported to have limited performance on prediction tasks, by only considering disease hierarchical structures, while neglecting disease horizontal links that reflect hidden disease complications [24]. Recent studies [23, 24] considered more complicated or dynamic information such as relationships between patients and diseases, or disease transition processes. Despite moderate progress, existing systems typically considered certain types of information. This makes it difficult for practitioners to decide which information should be used for various scenarios. Consequently, several challenges remain in effectively utilizing various information for patient similarity learning.
Q1: How to effectively utilize multi-modal information?
Q2: How to effectively incorporate the domain knowledge of medical concepts?
Q3: How to collaboratively learn hidden interactions from various information?
We propose a novel graph representation learning framework, denoted by One-Size-Fits-Three (OSFT), to uniformly support patient similarity learning by using various information. We first construct a global medical knowledge graph using the historical EHRs of all patients. Then, we build a heterogeneous graph for each patient to integrate local and external information. To capture hidden correlations, we propose a graph representation learning framework. Our OSFT framework mainly consists of three modules: node embedding, local and global interaction learning. We incorporate medical ontology and multi-modal data into learning node features for capturing hidden correlations and use a fusion-attention network to give different weights to different features to obtain more prepared node embeddings. For a patient, we feed its node embeddings and graph structure into two interaction learning modules. To learn a local representation, a neighbor-attention network is used to give different neighbors with different weights to capture the heterogeneous neighbor information. While a global-attention network is proposed to learn different weights for different nodes and aggregate node embeddings into a global graph representation. Finally, both global and local representations are fed into a fully connected layer to predict the final similarity score between two patients. Our main contributions are summarized as follows:
-
We propose a novel heterogeneous graph framework to represent a patient from EHRs by incorporating internal and external information. The constructed medical knowledge graph and heterogeneous patient graphs will be published for future research to do bench-marking evaluation.
-
We propose OSFT, to simultaneously learn patient representations and pairwise similarity. Three attention mechanisms are used to capture interaction signals from various information for generating more informative patient representations. This is the first effort to propose a generic framework that can uniformly support patient similarity learning with various information.
-
The results on two public datasets verify the effectiveness and generality of our framework. When compared with baselines in the task of patient similarity search, OSFT achieves superior effectiveness and comparative efficiency. We also give new insights that whether the use of various information or graph model can better learn the patient similarity from EHRs.
2 Related Work
2.1 Patient Similarity
Patient Similarity Search Early methods applied traditional information retrieval techniques to patient similarity search [35]. Most of them retrieved medical events from EHRs and modeled them as feature vectors. The similarity among patients is computed as the distance between their medical event vectors. However, these methods do not perform well, due to the heterogeneous and sparse nature of EHRs. Recent studies connected medical concepts through hierarchical structures and employed deep learning models for patient similarity learning [15].
Patient Similarity Learning Existing work have three groups by using various information: multi-modal EHRs, external knowledge and correlations among medical concepts. The first group adopted multi-modal EHRs to learn patient representations [14, 20, 29, 36]. Early work considered textual or visual similarities between EHRs [14, 29]. Later work employed multi-modal information for representation learning [20, 36]. For example, Li et al. [20] presented a self-supervised feature learning method using multi-modal EHRs. However, due to the sparse nature of EHRs, these methods suffered from the data insufficiency problem [27]. The second group incorporated external information, such as medical concept descriptions [26] and ontology [6, 22, 27, 30, 34], into learning the embeddings. GRAM [6] supplemented EHRs with hierarchical information from medical ontology. Lin et al. [22] used a medical knowledge graph to learn the embeddings of medical entities. D3K [30] used domain knowledge and data-driven insights for patient similarity measurement. However, external sources may be limited in terms of quantity and quality [8]. The third group captured hidden information such as correlations among medical concepts. Early work embedded medical concepts using one-hot representations [2]. Several methods incorporated semantic information into embedding medical concepts [5, 8, 10]. MiME [8] modeled the EHRs as hierarchical graphs and used neural networks to learn the patient representations. However, EHRs do not always contain complete structure information. Thus, GCT [9] jointly learned the hidden structure of EHRs while performing various prediction tasks when the structure information is unavailable. Several work attempted to learn the medical concept embeddings using temporal information in EHRs by adopting existing neural networks [7, 8, 25, 41]. Retain [7] proposed a two-level attention model to learn the clinical representations. Dipole [25] learned the embeddings with bidirectional recurrent neural networks. Deep Embedding [41] employed the convolutional neural network to learn the patient representations. However, they often required big data volume of EHRs for learning effective embeddings.
We observe that: (1) Recent studies that employ graph neural networks to learn patient representations have been reported to have state-of-the-art performance. (2) Due to the heterogeneous and sparse nature of EHRs, patient graphs constructed based on EHRs are sparser than general heterogeneous graphs, which requires the patient similarity task to model EHRs in a more accurate manner.
Based on these observations, we propose an effective framework to uniformly support patient representation learning using various information, including multi-modal EHRs, external knowledge, and correlations among medical concepts. In this way, the heterogeneous graph we build for each patient integrates local and external information in the nodes, which are different from the general heterogeneous graph. A graph learning framework is also developed to learn interaction signals among various information for accurately measuring patient similarity.
2.2 Graph Neural Networks
Graph Neural Networks (GNNs) are widely explored to process graph data. GGNN [21] is a broadly used model that has favorable inductive biases relative to purely sequence-based models when the problem is graph-structured. GCN [19] is a semi-supervised learning approach on graph data that is based on an efficient variant of convolutional neural networks which operate directly on graphs. GAT [37] leveraged masked self-attention layers to capture hidden information based on graph convolutions or their approximations. Xu et al. [39] developed a simple neural architecture, denoted as Graph Isomorphism Network (GIN), and showed that its representation power is equal to the power of the Weisfeiler-Lehman test.
This paper employs GNNs to capture interaction information among various information for patient similarity learning. We also conduct an ablation study to evaluate the effectiveness of various GNNs in Sect. 5.4.
3 The Heterogeneous Patient Graph
This paper first builds a medical knowledge graph from EHRs as follows.
Definition 1
Medical Knowledge Graph. The medical knowledge graph MKG is constructed from EHRs, which contains three types of entities: a diagnosis denoted as \(v^d\), a treatment denoted as \(v^t\) and a medicine denoted as \(v^m\). The diagnoses and treatments are mapped to the medical codes of International Classification of Disease (ICD-9) [32], and the medicines are obtained from the National Drug Code.Footnote 1 There are two types of relationships, denoted as triples of \((v^d, r^m, v^m)\) and \((v^d,r^t,v^t)\), where \(r^m\) represents the relationship between a diagnosis and a medicine, and \(r^t\) represents the relationship between a diagnosis and a treatment. For example, the node of disease “Acute Myocardial Infarction” can be treated with the node of medication “Streptokinase” and the node of treatment "Coronary Bypass," so the relationship between “Acute Myocardial Infarction” node and “Streptokinase” node is \(r^m\) and the relationship between “Acute Myocardial Infarction” node and “Coronary Bypass” node is \(r^t\).
To extract the relationships from EHRs for constructing the MKG, this paper applies the co-occurrence information to connect the relationships among various medical concepts. We construct a representative graph for each patient using the medical knowledge graph as below:
Definition 2
Patient Graph. A patient graph is defined as \({\mathcal {G}}=\{{\mathcal {V}},{\mathcal {E}}\}\), where \({\mathcal {V}}\) is a finite set of nodes and \({\mathcal {E}}\) is a finite set of edges. We have \({\mathcal {V}}=\{{\mathcal {V}}^d \bigcup {\mathcal {V}}^t \bigcup {\mathcal {V}}^m \}\), where \({\mathcal {V}}^d = \{v^d_1,\dots ,v^d_{|{\mathcal {V}}^d|}\}\) is a set of diagnoses, \({\mathcal {V}}^t = \{v^t_1,\dots ,v^t_{|{\mathcal {V}}^t|}\}\) is a set of treatments, and \({\mathcal {V}}^m = \{v^m_1,\dots ,v^m_{|{\mathcal {V}}^m|}\}\) is a set of medicines. \(|{\mathcal {V}}^d|\), \(|{\mathcal {V}}^t|\) and \(|{\mathcal {V}}^m|\), respectively, represent the size of each set. We have \({\mathcal {E}}=\{{\mathcal {R}}^m \bigcup {\mathcal {R}}^t\}\). \({\mathcal {R}}^m = \{r^m_1,\dots ,r^m_{|{\mathcal {R}}^m|}\}\) is a set of relationships among diagnoses and medicines. \({\mathcal {R}}^t = \{r^t_1,\dots ,r^t_{|{\mathcal {R}}^t|}\}\) is a set of relationships among diagnoses and treatments.
Given the EHRs of a patient, we first extract three types of entities from the EHRs based on the medical knowledge graph of MKG, which are used to construct the nodes of the patient graph. Then, given each candidate pair of nodes, if there exists a relationship between their corresponding entities in the MKG, we add an edge between these two nodes in the patient graph. Here, a candidate pair of nodes should be nodes of diagnoses and medicines, or that of diagnoses and treatments. To enhance the representation ability, we propose a novel structure, denoted as a heterogeneous patient graph, for incorporating external information into the patient graph.
Definition 3
Heterogeneous Patient Graph. Given a patient graph of \({\mathcal {G}}=\{{\mathcal {V}},{\mathcal {E}}\}\), we attach external sources into its nodes to construct the heterogeneous patient graph. A heterogeneous patient graph is defined as a 4-tuple of \(G=\{V,E,\varSigma ,{\mathcal {L}}\}\), where V is a finite set of attributed nodes, \(E \subseteq V \times V\) is a finite set of edges, \(\varSigma\) is a finite set of attribute sets and \({\mathcal {L}}:V \rightarrow \varSigma\) is a mapping function assigning a set of attributes to a node. Here, an attribute set is defined as a set of external information sources including a medical image, a textual note and a medical ontology.
In general, V is obtained from \({\mathcal {V}}\) by attaching each node in \({\mathcal {V}}\) with a set of attributes. For nodes of diagnoses and treatments, we use their medical concepts as keywords to retrieve their medical images from Google imagesFootnote 2 and MedPixFootnote 3; while for those of medicines, we collect their images from IndiaMart.Footnote 4 A textual note can be obtained from doctor notes in EHRs, or medical concept descriptions. The medical ontology is represented as a directed acyclic graph (DAG) [11]). For nodes of diagnoses and treatments, we obtain their medical ontology from the ICD-9 ontology [32]; while for those of medicines, we obtain their medical ontology from the ATC ontology [3]. The relationships in E are directly inheriting from those in \({\mathcal {E}}\).
Figure 1 illustrates an example of our constructed heterogeneous patient graph. The various colors represent various types of medical concepts. Each node is attached with external data. For example, the node of diagnose “Acute Myocardial Infarction” is attached with a medical image, a textual note and the medical ontology partially extracted from the ICD-9 ontology.
Definition 4
Problem Statement. Given a pair of patients, we first construct two heterogeneous patient graphs of \(G_1\) and \(G_2\) for them. The objective is to calculate the similarity score, denoted as \(S(G_1,G_2)\), between the pairwise graphs. We normalize the value of \(S(G_1,G_2)\) into a range of [0, 1] and classify two patients as similar ones if the value of \(S(G_1,G_2)\) is greater than a given threshold value and vice versa. In this paper, the default threshold value is set to 0.5 without loss of generality.
4 The Generic Learning Framework
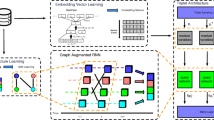
Given two heterogeneous patient graphs, we develop a novel graph representation learning framework, denoted by One-Size-Fits-Three (OSFT), to uniformly support the patient similarity learning using various information. Figure 2 gives an overview of our approach. Our OSFT contains three main modules: node embedding, local interaction learning and global interaction learning. For node embedding, we incorporate the attributes of each node into learning the embedding. For each patient, we feed its node embeddings and heterogeneous graph into the local and global interaction learning modules, respectively, to learn its local and global representations, which are finally fed into a fully connected layer to predict the final similarity score between two given patients.
The proposed OSFT framework. Given two patient graphs, we first generate node embeddings using our proposed multi-modal representation learning method. The node embeddings are then fed into the graph similarity learning component as initial embeddings. Then, the node embeddings are updated by employing multiple GGNN layers. Based on the initial and updated embeddings, two interaction learning strategies are adopted to capture the global and local interaction signals, respectively. Finally, both interaction signals are fed into a fully connected layer to predict the graph similarity score between the pairwise graphs
4.1 Node Embedding
Given a heterogeneous patient graph of \(G=\{V,E,\varSigma ,{\mathcal {L}}\}\), we encode each node \(v_n^* \in V\) into an embedding of \({\varvec{m}} \in {\mathbb {R}}^{d_m}\), where \(d_m\) is the dimension. We propose a generic node embedding module to encode various information for the node. The image encoder and text encoder are employed to generate the image embedding \({\varvec{e}} \in {\mathbb {R}}^{d_e}\) and text embedding \({\varvec{t}} \in {\mathbb {R}}^{d_t}\) separately. Without loss of generality, we generate the image embedding of 2, 048 dimensions using the pre-trained ResNet50 [13], and the text embedding of 300 dimensions using Word2Vec [10].
This paper incorporates external medical ontology into the node embedding. As described in Sect. 3, the medical ontology is modeled as a DAG. We derive the ontology embedding \({\varvec{o}} \in {\mathbb {R}}^{d_o}\) for the node \(v_n^*\) based on its corresponding leaf node in the DAG. We first initialize the medical code of \(v_n^*\), denoted as \(c_*\), in the DAG with a vector \({\varvec{r}}_* \in {\mathbb {R}}^{m}\). Then, we employ two strategies with GAT [37] to generate the final embedding.
Given a DAG, we employ a bottom-to-top strategy to obtain the embedding \({\varvec{h}}_i \in {\mathbb {R}}^{m}\) of each node \(c_i\) in the DAG by integrating the information from its children denoted by \(ch(c_i)\) as follows.
Here, \({\varvec{W}} \in {\mathbb {R}}^{m\times m}\) is a weight matrix for input transformation, \({\varvec{a}} \in {\mathbb {R}}^{2m}\) is a weight vector, and \(LeakyReLU(\cdot )\) is a nonlinear function.
Then, we use an ancestor-to-leaf strategy to generate the embedding \({\varvec{h'_{i'}}} \in {\mathbb {R}}^{d_o}\) for each leaf node \(c_{i'}\) in the DAG by combining all embeddings from its ancestors \(anc(c_{i'})\).
Here, \({\varvec{W'}} \in {\mathbb {R}}^{d_o\times m}\) and \({\varvec{a'}} \in {\mathbb {R}}^{2d_o}\) are the learnable parameters similar to \({\varvec{W}}\) and \({\varvec{a}}\) in Eq. 1.
4.1.1 Fusion Mechanisms
To fuse the obtained embeddings into an effective node embedding, this module also supports various fusion mechanisms. We show one mechanism as an example for space limits. We normalize the image embedding \({\varvec{e}}\), text embedding \({\varvec{t}}\) and ontology embedding \({\varvec{o}}\) by projecting them into a common space to get \({\varvec{e'}} \in {\mathbb {R}}^d\), \({\varvec{t'}} \in {\mathbb {R}}^d\) and \({\varvec{o'}} \in {\mathbb {R}}^d\). We then concatenate the projected embeddings to generate the multi-modal embedding \({\varvec{m}} \in {\mathbb {R}}^{3d}\) for each node as: \({\varvec{m}}= {{\varvec{[}}e';\,t';\,o'}]\), where [; ] is the concatenation operator.
4.2 Patient Similarity Learning
Given two patients with \(G_1\) and \(G_2\), the derived node embeddings for them are \({\varvec{M_1}} \in {\mathbb {R}}^{|V_1|\times {3d}}\) and \({\varvec{M_2}} \in {\mathbb {R}}^{|V_2|\times {3d}}\), respectively, where \(|V_1|\) and \(|V_2|\) are the number of nodes in two patient graphs. Based on the input node embeddings, we employ various graph neural networks such as GAT, GGNN, GCN and GIN, to update the embedding of each node by capturing the structural information. From the experimental results in Table 6, we find the GGNN showing better performance. We use L layers of GGNN to update the node embeddings. The updating of the hidden representation for a node \(v^*_n\) are performed as GRU [4] with the initial state of \({\varvec{h^0_{n}}} = [{\varvec{m_n}};\,{\textbf{0}}]\), where \({\varvec{m_n}}\) is the input node embedding of \(v^*_n\). At the \({(l+1)}\)-th layer, the transition from \({\varvec{h^{l}_n}} \in {\mathbb {R}}^{3d}\) to \({\varvec{h^{l+1}_n}} \in {\mathbb {R}}^{3d}\) can be formulated as:
Here, \({\mathcal {N}}(n)\) is the neighbors of node \(v^*_n\), \({\varvec{a^{l+1}_n}}\) is an aggregated hidden representation from nodes \({\mathcal {N}}(n)\), \({\varvec{z^{l+1}_n}}\) and \({\varvec{r^{l+1}_n}}\)i, which indicate the update vector and reset gate vector. \(\sigma (\cdot )\) is the activation function. \({\varvec{W_z}}\), \({\varvec{W_r}}\), \(\widetilde{{\varvec{W}}}\), \({\varvec{b_z}}\), \({\varvec{b_r}}\) and \(\widetilde{{\varvec{b}}}\) are the common parameters used as the GRU. The hidden state of each node is updated dynamically with the information from its neighbors and previous GGNN layer. Thus, both the original representation and structural information of the node can be preserved.
4.2.1 Global Interaction Learning
We propose an attention mechanism to learn the node weights and aggregate the node embeddings into a graph representation. Based on the graph representation, we further utilize the neural tensor network following [33] to capture the global interaction information from multiple scales.
Taking \(G_1\) as an example, we first extract the embedding \({\varvec{h}}^{L}_n\) learned from the last GGNN layer and the input node embedding \({\varvec{m}}_n\) for node \(v^*_n\). Different from the initial embedding \({\varvec{m}}_n\), the updated embedding \({\varvec{h}}^{L}_n\) incorporates the complex structural information of \(v^*_n\). If the difference is bigger, \(v^*_n\) may retain more complex connections with other graph nodes and should receive a higher attention weight. Thus, the graph embedding \({\varvec{g}}_1 \in {\mathbb {R}}^{3d}\) learned from the specific attention mechanism can be formulated as:
where \(sigmoid(({\varvec{m}}_n{\varvec{w}})^{T}{\varvec{h}}^L_n)\) calculates the attention weight for node \(v^*_n\), with \({\varvec{w}} \in {\mathbb {R}}^{3d}\) as the learnable parameters.
Akin to \(G_1\), we also utilize the attention mechanism to obtain the graph representation \({\varvec{g_2}}\) for \(G_2\). Inspired by the strengths of Neural Tensor Networks [33] in modeling the relation between two vectors, we employ it to capture the global interaction signals from K scales as \({\varvec{S_1(G_1,G_2)}}\):
where \({\varvec{W_4^{[1:K]}}} \in {\mathbb {R}}^{3d \times 3d \times K}\) and \({\varvec{W_5}} \in {\mathbb {R}}^{K \times 6d}\) are weight matrices, and \({\varvec{b_4}} \in {\mathbb {R}}^K\) is a bias vector. K is a hyperparameter controlling the number of interaction (similarity) scores produced by the model for each graph embedding pair.
4.2.2 Local Interaction Learning
We propose a local interaction learning strategy to capture local important information for learning the patient similarity. We first calculate the multi-level local interaction signals based on different levels of node embeddings. We perform the local interaction operation on the diagnose nodes which are recognized as the most important healthcare information [9]. Given the diagnose node \(v^d_i \in G_1\) and \(v^d_j \in G_2\), we use the cosine similarity to measure the multi-level interaction signals from the input node embeddings and L updated node embeddings.
We calculate the multi-level interaction signals between each node pairs and build interaction matrices \(\{{\varvec{P^0}},{\varvec{P^1}},\dots ,{\varvec{P^L}}\}\). We use the matching histogram mapping of \(d_q\) bins to vectorize the matrices into multi-level matching patterns \(\{{\varvec{q^0}}, {\varvec{q^1}},\dots , {\varvec{q^L}}\} \in {\mathbb {R}}^{d_q}\) following [12]. The size of our local interactions is not fixed due to the variable node sizes. Therefore, we can generate the high-level local information without considering the variable node sizes and truncating the nodes. It groups local interactions into a set of ordered bins according to different levels of matching strengths. We apply the logarithm over the grouped value in each bin as the hierarchical matching pattern. Finally, we obtain the multi-level local interaction signals \({\varvec{S_2(G_1,G_2)}}\) as below:
Finally, \({\varvec{S_1(G_1,G_2)}}\) and \({\varvec{S_2(G_1,G_2)}}\) are fed into a fully connected neural network to compute the final similarity score \({\varvec{S(G_1,G_2)}}\). We use the mean square error loss as the objective function to be minimized. For two similar patients, the ground truth similarity score \(S_{gt}(G_1,G_2)\) is 1, while that of dissimilar ones is 0. The loss is computed as:
where \({\mathcal {D}}\) is the training graph pairs (Table 1).
5 Experiments
We aim to answer the following four questions:
-
Does our OSFT outperform strong baselines on the patient similarity learning task?
-
Does our OSFT perform efficiently?
-
Does external information contribute to our learning task?
-
What is the effect of global or local learning modules?
5.1 Setup
MIMIC-III [17] and MIMIC-IV [16] are two real-world EHRs datasets. MIMIC-III is obtained from over 40, 000 patients who are admitted to hospitals from 2001 to 2012, while MIMIC-IV includes the data of admission to the intensive care unit from 2008 to 2019.
We construct a knowledge graph using MIMIC-III and MIMIC-IV datasets, and the specific statistical information of MIMIC-III and MIMIC-IV involved in the experiments is shown in Table 2. We divide each dataset into a training set (60\(\%\)), a test set (20\(\%\)) and a validation set (20\(\%\)). Each patient contains one diagnose code and belongs to only one cohort. We convert all the patients to the patient graphs based on knowledge graphs. Given a patient, we randomly pick several patients from its cohort and label each pair of patients as similar with a ground truth similarity score of 1. We also randomly pick several patients from other cohorts to form dissimilar pairs with a score of 0. The statistics are in Table 1. We classify two patients into a similar and dissimilar pair and evaluate the results using two popular metrics: Accuracy and F1 score.
Our baselines include: (1) five state-of-the-art models for patient similarity measuring including Auto-Diagnosis [38], MiME [8], GCT [9], GRAM [6] and Deep Embedding [41]; (2) two adapted methods from graph matching algorithms including Graph2vec [28] and SimGNN [1].
Our OSFT is implemented with PytorchFootnote 5 and trained using Adam optimizer [18] with the learning rate of \(5\textrm{e}{-4}\). We trained our model for 100 epochs with the batch size of 256. The dimension of node embeddings is 100 and the number of the GGNN layer is 1. We set K in the neural tensor network and the number of bins in the matching histogram mapping both to 20. For baselines, we set the threshold value in Auto-Diagnosis to 50. The learning rate and embedding size for Deep Embedding are 0.001 and 300, respectively. We implemented GCT with 3 layers and trained it for 1000 epochs with a learning rate of 0.001. We trained GRAM and MiME with the initial learning rate of \(5\textrm{e}{-4}\) for 200 epochs. For Graph2vec, we use the default parameters in Doc2Vec implemented with gensim.Footnote 6 For SimGNN, we trained it for 50 epochs with a learning rate of 0.001.
5.2 Performance
In Table 3, our OSFT consistently achieves the best performance on all metrics. This suggests that our graph framework indeed enhances learning the patient similarity. The results of deep learning methods including MiME, GRAM, Graph2vec, Deep Embedding, SimGNN, GCT and our OSFT steadily outperform traditional methods (i.e., Auto-Diagnosis). The methods considering external information, such as our OSFT, GCT and SimGNN, achieve better performance. The method based on temporal patient representations (i.e., Deep Embedding) outperforms the methods using static information such as MiME, GRAM and Graph2vec. This suggests that incorporating dynamic structure information or external knowledge can enhance patient similarity learning.
5.3 Efficiency
Figure 3 shows the results on training time. No training is required for Auto-Diagnosis and Graph2vec. GRAM and MiME take the smallest training time, as they only consider static information on EHRs. However, they show very poor performance. Our OSFT requires comparative training time. Figure 4 presents the average time required to predict a pair of patients on MIMIC-III. Due to the limitation of the space, we omit the similar results on MIMIC-IV. The results show that our OSFT requires comparative prediction time while achieving the best performance.
5.4 Ablation Study
We conduct an ablation study by eliminating the text (w/o text embedding), image (w/o image embedding) or ontology sources (w/o ontology embedding) from our framework. We also evaluate the effect of global (w/o global signals) or local learning module (w/o local signals). We omit the similar results of various fusion mechanisms (Sect. 4.1.1) for space limit. In Table 4, incorporating external information indeed contributes to the task, and the text embedding boost the performance most significantly. This indicates that the textual information extracted from clinical notes is important for patient similarity learning. The model that integrates only one form of information can achieve competitive results with baselines. This indicates that external sources can enrich patient representations.
As shown in Table 4, removing any kind of interaction signals will lead to poor performance on both metrics, which means that both factors contribute to enhancing patient similarity search. Global interaction learning considers the patient’s overall situation, while local interaction learning considers some specific details of the patient’s treatment and medication, so both of them are useful for the patient similarity search task. On the other hand, OSFT assigns higher importance to local interaction signals, proving the necessity of using useful node-level interaction information. The global interaction signals are derived from using Neural Tensor Networks to model two graph representation vectors, but graph embedding may lose local structural information. Whereas local interaction signals contain high-level local information of all nodes without considering node size changes and truncated nodes, which is an important reason why the local interaction signals exhibit a more powerful effect than the global interaction signals.
We evaluate the effect of various fusion mechanisms in our framework, and the results are shown in Table 5. The Concat Attention shows a slight performance advantage over the fusion method of concatenation and addition. We also evaluate the effect of various graph neural networks and attention mechanisms in our framework, and the results are shown in Table 6. Various GNNs show slight difference on the performance, which indicates that node aggregation methods did not affect our similarity learning results. The reason may be that our framework can mostly capture hidden information, and this helps to reduce the affection from node aggregation functions. The framework with the global cross attention outperforms that with the simple softmax. This indicates that incorporating external information requires the attention to avoid introducing too much noisy information.
5.5 Sensitivity Testing
Figure 5 shows the sensitivity testing on MIMIC-III. In Fig. 5a, the performance is stable when the dimension is less than 200, while it drops when the dimension becomes larger than 200. Therefore, we set the dimension to 100 in our experiments. Figure 5b shows the performance decreases gradually when the number of GGNN layers increases. This indicates that one-hop relationship is the most effective information for enhancing a node embedding in our task. Thus, we set the number of GGNN layers to 1. In Fig. 5c, the performance first increases as the number of bins becomes larger, while decreases when the number becomes larger than 20. Therefore, we set the number of bins used in our local interaction learning module as 20.
5.6 Case Study
We rank similar patients from a candidate set based on our similarity scores. The results are shown in Table 7. Given a query patient (“121885”) having a disease of “Coronary atherosclerosis of native coronary artery,” the candidate set contains three similar patients (“156013,” “139873” and “122310”) having the same disease, two similar patients (“199137,” “175817”) suffering from a disease in the same cohort as that of the query patient, and five dissimilar patients randomly selected from other cohorts. The patients returned by our OSFT are: three patients having the same disease are ranked as the top-3 results and one patient having a similar disease is ranked as the \(4^{th}\) result. Compared with baselines, our OSFT achieves the best results. SimGNN and Auto-Diagnosis return four patients having the same or similar diseases with error ranking orders. MiME returns three patients having the same disease while missing all patients with similar diseases. Others perform even worse.
6 Conclusion
We propose a novel graph structure to model EHRs of patients by effectively incorporating external information sources. A novel generic learning framework called by OSFT is proposed to uniformly support the patient similarity learning by using various information. Our OSFT framework can capture both the global graph-level and local node-level interaction information for learning the similarity scores between two patients. The experimental results verify the generality and effectiveness of our framework. Our framework achieves superior effectiveness and comparative efficiency, compared to strong baselines. The results also provide new insights about whether the use of various information can better measure the patient similarity.
Data Availability
Source codes and datasets: https://github.com/emmali808/ADDS/tree/master/EHRDeepHelper.
References
Bai Y, Ding H, Bian S, Chen T, Sun Y, Wang W (2019) Simgnn: a neural network approach to fast graph similarity computation. In: WSDM, pp. 384–392
Chen R, Su H, Khalilia M, Lin S, Peng Y, Davis T, Hirsh DA, Searles E, Tejedor-Sojo J, Thompson M, et al (2015) Cloud-based predictive modeling system and its application to asthma readmission prediction. In: AMIA Annual Symposium Proceedings. American Medical Informatics Association, vol. 2015, p. 406
Cheng X, Zhao SG, Xiao X, Chou KC (2017) iatc-mhyb: a hybrid multi-label classifier for predicting the classification of anatomical therapeutic chemicals. Oncotarget 8(35):58494
Cho K, Van Merriënboer B, Bahdanau D, Bengio Y (2015) On the properties of neural machine translation: encoder-decoder approaches. arXiv preprint arXiv:1409.1259
Choi E, Bahadori MT, Searles E, Coffey C, Thompson M, Bost J, Tejedor-Sojo J, Sun J (2016) Multi-layer representation learning for medical concepts. In: SIGKDD, pp. 1495–1504
Choi E, Bahadori MT, Song L, Stewart WF, Sun J (2017) Gram: graph-based attention model for healthcare representation learning. In: SIGKDD, pp. 787–795
Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W (2016) Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. NIPS 2(9):3504–3512
Choi E, Xiao C, Stewart W, Sun J (2018) Mime: multilevel medical embedding of electronic health records for predictive healthcare. In: NIPS, pp. 4547–4557
Choi E, Xu Z, Li Y, Dusenberry M, Flores G, Xue E, Dai A (2020) Learning the graphical structure of electronic health records with graph convolutional transformer. AAAI 34:606–613
Choi Y, Chiu CYI, Sontag D (2016) Learning low-dimensional representations of medical concepts. AMIA Summits Transl Sci Proceed 2016:41
Christofides N (1975) Graph theory: an algorithmic approach (Computer science and applied mathematics). Academic Press, Cambridge
Guo J, Fan Y, Ai Q, Croft WB (2016) A deep relevance matching model for ad-hoc retrieval. In: CIKM, pp. 55–64
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: CVPR, pp. 770–778
He Z, Yang J, Wang Q, Li J (2016) A method of electronic medical record similarity computation. In: ICSH, pp. 182–191. Springer
Huang HZ, Lu X, Guo W, Jiang XB, Yan ZM, Wang SP (2021) Heterogeneous information network-based patient similarity search. Front Cell Develop Biol 9:735687
Johnson A, Bulgarelli L, Pollard T, Horng S, Celi LA, Roger M (2021) MIMIC-IV (version 1.0) https://doi.org/10.13026/s6n6-xd98
Johnson AE, Pollard TJ, Shen L, Li-Wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG (2016) Mimic-iii, a freely accessible critical care database. Scient. Data 3(1):1–9
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Kipf T, Welling M (2017) Semi-supervised classification with graph convolutional networks. arXiv:1609.02907
Li X, Jia MJ, Islam MT, Yu L, Xing L (2020) Self-supervised feature learning via exploiting multi-modal data for retinal disease diagnosis. TMI 3(9):4023–4033
Li Y, Tarlow D, Brockschmidt M, Zemel R (2015) Gated graph sequence neural networks. arXiv preprint arXiv:1511.05493
Lin Z, Yang D, Jiang H, Yin H (2021) Learning patient similarity via heterogeneous medical knowledge graph embedding. IJCS, 8(4)
Lu C, Han T, Ning Y (2022) Context-aware health event prediction via transition functions on dynamic disease graphs. AAAI 36:4567–4574
Lu C, Reddy CK, Chakraborty P, Kleinberg S, Ning Y (2021) Collaborative graph learning with auxiliary text for temporal event prediction in healthcare. In: IJCAI, pp. 3529–3535
Ma F, Chitta R, Zhou J, You Q, Sun T, Gao J (2017) Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In: SIGKDD, pp. 1903–1911
Ma F, Wang Y, Xiao H, Yuan Y, Chitta R, Zhou J, Gao J (2019) Incorporating medical code descriptions for diagnosis prediction in healthcare. BMC Med Inform Dec Mak 19(6):1–13
Ma F, You Q, Xiao H, Chitta R, Zhou J, Gao J (2018) Kame: knowledge-based attention model for diagnosis prediction in healthcare. In: CIKM, pp. 743–752
Narayanan A, Chandramohan M, Venkatesan R, Chen L, Liu Y, Jaiswal S (2017) graph2vec: Learning distributed representations of graphs. arXiv preprint arXiv:1707.05005
Ni J, Liu J, Zhang C, Ye D, Ma Z (2017) Fine-grained patient similarity measuring using deep metric learning. In: CIKM, pp. 1189–1198
Oei RW, Fang HSA, Tan WY, Hsu W, Lee ML, Tan NC (2021) Using domain knowledge and data-driven insights for patient similarity analytics. JPM 1:1
Parimbelli E, Marini S, Sacchi L, Bellazzi R (2018) Patient similarity for precision medicine: A systematic review. JBI 8(3):87–96
Slee VN (1978) The international classification of diseases: ninth revision (icd-9)
Socher R, Chen D, Manning CD, Ng A (2013) Reasoning with neural tensor networks for knowledge base completion. NIPS 2(6):926–934
Song L, Cheong CW, Yin K, Cheung WK, Fung BC, Poon J (2019) Medical concept embedding with multiple ontological representations. In: IJCAI, pp. 4613–4619
Suo Q, Zhong W, Ma F, Yuan Y, Huai M, Zhang A (2018) Multi-task sparse metric learning for monitoring patient similarity progression. ICDM pp. 477–486
Jiménez-del Toro OA, Otálora S, Atzori M, Müller H (2017) Deep multimodal case-based retrieval for large histopathology datasets. In: International Workshop on Patch-MI, pp. 149–157. Springer
Velickovic P, Cucurull G, Casanova A, Romero A, Lio’ P, Bengio Y (2018) Graph attention networks. arXiv:1710.10903
Wang X, Wang Y, Gao C, Lin K, Li Y (2018) Automatic diagnosis with efficient medical case searching based on evolving graphs. IEEE Access 6:53307–53318
Xu K, Hu W, Leskovec J, Jegelka S (2019) How powerful are graph neural networks? arXiv:1810.00826
Zhou X, Li Y, Liang W (2020) Cnn-rnn based intelligent recommendation for online medical pre-diagnosis support. TCBB pp. 912–921
Zhu Z, Yin C, Qian B, Cheng Y, Wei J, Wang F(2016) Measuring patient similarities via a deep architecture with medical concept embedding. IEEE, In: ICDM, pp. 749–758
Acknowledgements
We acknowledge the editorial committee’s support and all anonymous reviewers for their insightful comments and suggestions, which improved the content and presentation of this manuscript.
Funding
Xiaoli Wang was supported by the Natural Science Foundation of Fujian Province of China (No. 2021J01003). Bin Luo was funded by the Central Guidance on Local Science and Technology Development Funds.
Author information
Authors and Affiliations
Contributions
Material preparation, data collection and analysis were performed by YH, FL. The first draft of the manuscript was written.
Corresponding author
Ethics declarations
Conflcit of ineterest
The authors declare that they have no confict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, Y., Luo, F., Wang, X. et al. A One-Size-Fits-Three Representation Learning Framework for Patient Similarity Search. Data Sci. Eng. 8, 306–317 (2023). https://doi.org/10.1007/s41019-023-00216-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41019-023-00216-9