Abstract
Phytopathometry can be defined as the branch of plant pathology (phytopathology) that is concerned with estimation or measurement of the amount of plant disease expressed by symptoms of disease or signs of a pathogen on a single or group of specimens. Phytopathometry is critical for many reasons, including analyzing yield loss due to disease, breeding for disease resistance, evaluating and comparing disease control methods, understanding coevolution, and studying disease epidemiology and pathogen ecology. Phytopathometry underpins all activities in plant pathology and extends into related disciplines, such as agronomy, horticulture, and plant breeding. Considering this central role, phytopathometry warrants status as a formally recognized branch of plant pathology. The glossary defines terms and concepts used in phytopathometry based on disease symptoms or visible pathogen structures and includes those terms commonly used in the visual estimation of disease severity and sensor-based methods of disease measurement. Relevant terms from the intersecting disciplines of measurement science, statistics, psychophysics, robotics, and artificial intelligence are also included. In particular, a new, broader definition is proposed for “disease severity,” and the terms “disease measurement” and “disease estimate” are specifically defined. It is hoped that the glossary contributes to a more unified cross-discipline approach to research in, and application of the tools available to phytopathometry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The two synonymous terms “plant pathometry” or “phytopathometry” were first coined by Large (1953, 1966). Phytopathometry as defined by Nutter et al. (1991) equates with “disease assessment” and is the branch of the discipline of phytopathology that deals with estimation or measurement of the amount of plant disease (broadly encompassing detection, identification, and quantification). Large (1953) stated, “At this point what we have been calling ‘disease measurement’ or ‘disease assessment’ enters a new phase. It becomes a systematized and specialized method of mensuration with a derived superstructure. In short, it becomes a branch of plant pathological science, and I think it deserves a better name. The name that I would now, and hereby, propose for it is ‘Plant Pathometry’, from pathos, disease or suffering, and metron, measure.” In the article, Large provides examples (including late blight of potato and choke of cocksfoot) where the new science of phytopathometry was a basis for obtaining valuable information from plant disease surveys. Thus, phytopathometry may be considered the branch of the discipline of phytopathology concerned with the detection, identification, and quantification of disease symptoms or signs of a pathogen.
Large (1966) stated and Gregory (1982) reiterated five requirements of disease measurement in relation to yield loss analyses: (1) a description of the morphology and development of the healthy crop; (2) a study of the course of the disease on plants in the field; (3) preparing standard area diagrams for detailed assessment of disease intensity, followed later by a simplified field assessment key (or field key) for field use (a field key was defined as an aid for the rapid visual assessment of leaf disease on whole plants or plots, or specific sampling areas (Large 1966)); (4) conducting field trials over a number of years, assessing disease progress using a field key for the disease assessments, and recording yields of plots with uncontrolled infection compared with plots kept free from disease; and finally (5) use of the disease progress curves to select particular assessment points (host growth stages) that will define severity in relation to loss of yield.
Most certainly, requirements 1 to 3 are foundational to phytopathometry (the healthy state must be known, ranges in disease symptoms understood, and diagrams, field keys, or other methods developed to aid assessments). However, requirements 4 and 5 are related to the analysis of the disease assessment results specifically in relation to crop loss assessment. Disease intensity data are used for epidemiological studies and are the basis for deriving yield loss models. The tactical view of phytopathometry described by Large (1966) and reiterated by Gregory (1982) could be considered an expanded view as it encompasses yield loss. However, Large (1966) also states, “Disease measurement is often regarded as a synonym for ‘estimation of losses,’ but this is misleading.” And Large further states, “The main purpose of work on disease measurement is to improve all plant disease recording or reporting, by making it not only qualitative but also much more quantitative.” Furthermore, plant disease quantification is vital for a myriad of reasons, including understanding the impact of disease on yield, breeding for plant disease resistance, evaluating and comparing disease control methods, understanding coevolution of plant and pathogen populations, and studying disease epidemiology and pathogen ecology (Madden et al. 2007; Bock et al. 2010, 2016). It underpins all activities within our discipline and extends into related ones, such as agronomy, horticulture, and plant breeding. Sensu lato, phytopathometry might include measurement of the quantity of the pathogen in the host using molecular methods (but not, for example, measurements of the pathogen propagules in the air or soil), but we have chosen not to include molecular methods in this glossary, instead choosing to restrict the glossary to the more traditional definition of disease measurement based on symptoms or visible pathogen structures. Recently, a range of automated sensor-based digital technologies have been developed and are being adapted for use in phytopathometry (Bock et al. 2020; Mahlein 2016). Nonetheless, many of the same terms and concepts are identical and appropriate regardless of the methods used.
“Plant disease assessment” or phytopathometry is generally a section (within a chapter on another topic) in plant pathology textbooks as it is a key method used in many plant pathological studies (Agrios 2005; Lucas 2002; Tronsmo et al. 2020). Since ca. 1970, phytopathometry has been closely associated with botanical epidemiology given its strong quantitative component; and thus, phytopathometry has been a branch of plant pathology typically, but not exclusively, studied by plant disease epidemiologists. As with textbooks on plant pathology, plant disease assessment is commonly a chapter or section within reference or teaching texts on plant disease epidemiology (Cooke et al. 2006; Madden et al. 2007). Although epidemiologists have been at the forefront of the research related to phytopathometry, particularly related to the development and evaluation of methods, phytopathometry is, as noted, critical to many other plant pathology-related areas of science.
Based on the history of phytopathometry, its application, and the statements quoted from Large (1966), we contend that phytopathometry is concerned with the science of the measurement of plant disease. But phytopathometry is not subservient to the branches of epidemiology or yield loss, although it provides a critical service to these and other branches of plant pathology, as well as other disciplines. Thus, we aim to formulate this glossary to embrace phytopathometry as a distinct branch of plant pathology as Large (1953) suggested and that relates to all applications where disease is quantified (Fig. 1). We therefore envision phytopathometry at the overlapping intersection of plant pathology and measurement science but interfacing closely with three other research disciplines: imaging and sensing technology, psychophysics, statistics, and being an important resource to other disciplines that interface with phytopathology (Fig. 1).
Disease quantification has historically been performed visually or by means of an instrument, hence the use of the two terms: “estimate” (visual) or “measurement” (instrument or sensor based). The significant advances in both imaging and remote sensing technologies in the last two decades have directly impacted phytopathometry and its associated terms and concepts. Sensing technologies and image analysis have made substantial advances, and artificial intelligence has recently opened new horizons for phytopathometry, especially for obtaining objective measures of the quantity of plant disease. It has been 20 years since there was a glossary of terms and concepts used in phytopathometry (Nutter 2001), which was based on a similar sentinel work 10 years earlier (Nutter et al. 1991). The 1991 glossary was proposed after the first phase of quantitative research on plant disease assessment (1970 to 1990). It also presented concepts and terms from the perspective of quantifying crop loss and did not consider the many other purposes of disease quantification which are also critical. Thus, included herein are new terms (with respect to those defined in Nutter et al. (1991)), including many from imaging, sensing technology and artificial intelligence, and, more broadly, statistics and measurement science. We have also refined definitions or updated our understanding of some terms as used in plant disease assessment (Everitt 1998; Madden et al. 2007; Bock et al. 2010, 2016, 2020; Chiang et al. 2020). A call for enhanced dialogue between plant pathologists and remote sensing researchers was made recently (Heim et al. 2019), which is justified given the explosion in the number of applications of remote sensing and artificial intelligence to detect or measure plant diseases in the last 5 years (Bock et al. 2020). Indeed, the path to success in sensor-based phytopathometry is transdisciplinary research among plant pathologists, electrical engineers, agronomists, and informatics specialists (Mahlein 2016).
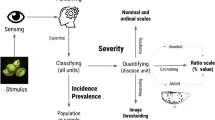
Before providing the updated and revised list of concepts and definitions, there are two terms that need to be clearly defined, with rationale, as they are so pivotal to phytopathometry and the practice of plant pathology. The first is “disease measurement.” Historically, this has referred to visual estimates of plant disease (Large 1966). However, with the advent of sensor-based technology, the disease can now be actually and accurately measured based on pixels or wave bands with healthy or diseased characteristics. Thus, we contend that disease measurement refers to those assessments made only using sensors and may refer to the number of individuals diseased, counts of a disease symptom on an organ, or the proportional quantity of disease on an individual specimen. In contrast, a “disease estimate” is one obtained through visual disease assessment, most commonly referring to estimates of the proportional quantity of disease on a specimen (but could be count-based data). The second term requiring discussion is “disease severity.” Here, we define disease severity as the “degree to which a specimen (plant or plant part) is diseased. Severity may be described quantitatively on a scale (for example, lesion length in mm, or area diseased in mm2) or as a proportion of the unit area diseased (commonly a percentage), as counts of the number of lesions present, or as a ranked numeric order (i) based on intervals of the percentage scale, or (ii) based on descriptions of symptoms in a progression from mild to severe (as applicable for many systemic diseases)” (Fig. 2). This definition differs from most prior definitions that described severity purely on the basis of the proportion of the specimen area diseased — but many plant diseases show degrees of severity without an easily defined area affected (as noted, many systemic diseases). Thus, severity is broadened to be applicable to almost all plant diseases where it can be rated quantitatively using ordinal or ratio scales, and not solely in the narrow sense of proportion (or percentage) of area diseased as defined by Nutter et al. (1991). Precedent also encourages the definition of severity to be revisited and broadened to provide a terminus communis to metrics of disease quantification on a specimen. Thus, Seem (1984) referred to disease density (i.e., lesions per leaf) as a form of severity, although McRoberts et al. (2003) acknowledged this was not the intended definition of Nutter et al. (1991), and decided to use density as a separate measure. We assert that lesion counts (or density) are indeed one of the metrics of severity. A perusal of the plant pathology literature will demonstrate that many authors already use these various metrics as measures of severity as described in numerous journal articles. For example, Pethybridge et al. (2020) and Cowling and Gilchrist (1980) referred to lesion counts as disease severity — the latter authors also used lesion size as a measure of severity. Quantitative ordinal scales based on intervals of the percentage scale to determine quantity of disease are well known and widely used, with the stated purpose of rating disease severity (Horsfall and Barratt 1945; Kousik et al. 2018; Urrea and Harveson 2014). Similarly, qualitative or descriptive ordinal scales are used for many diseases where diseased area estimates are not possible, including many systemic and viral diseases — in the literature, the rating has often been described as a disease severity metric (Gottwald et al. 1989; Ling and Scott 2007; Pascual et al. 2010; Cook et al. 2020; Wang et al. 2020). And there are many examples of the percentage scale being used to estimate disease severity, in accordance with the original and narrow definition of the previous studies (Nutter et al. 1991; Colson et al. 2003; Scherm et al. 2009; Bock et al. 2017). The fact that so many studies have used the term “severity” to describe the full range of metrics quantifying disease on an individual specimen suggests that the terms need redefining to allow plant pathologists to describe the quantity of disease on a specimen generically as “disease severity” without having to resort to semantic gymnastics.
The updated concept and definition of plant disease severity to encompass the original definition (percentage area diseased, Nutter et al. 1991), and the metrics of lesion counts or density, and ordinal scale-based measurements using classes based on either interval of the percentage scale or descriptions of symptoms as defined in this article
In addition, a review of the definitions for disease intensity, prevalence, and incidence (all of which remain unchanged) will augment the rationale for the case (for the relationships among these, see Fig. 3). We argue that broadening the term “disease severity” to include all metrics that can be used to rate a disease quantitatively using a numeric scale has many practical advantages. Finally, many terms here are defined specifically as they are used in phytopathometry and in some cases (e.g., incidence) may have different meanings in other disciplines.
The terms and concepts defined subsequently are from a broad range of sources, but most notably, they stem from those initially defined by Nutter et al. (1991) and from Nutter (2001), D'Arcy et al. (2001), Madden et al. (2007), Bock et al. (2010), and Bock et al. (2016). Other sentinel references are included (Everitt 1998; Nutter et al. 2006; McRoberts et al. 2003; Behmann et al. 2015; Del Ponte et al. 2017; Paulus and Mahlein 2020; Paulus 2019). The Special Topic article of Nutter et al. (1991) was the outcome of a subcommittee that was appointed by the Plant Disease Losses Committee of the American Phytopathological Society. The list of terms that we present in this glossary was compiled by the authors without specific societal authority but with the common purpose of bringing together in a single source, an updated and comprehensive (as of 2021) list of terms, definitions, and concepts currently used or making debuts in phytopathometry. This glossary includes 128 terms, which is an increment compared to the original glossary (Nutter et al. 1991) that had 58 terms and which included terms and concepts related to yield loss measurement. Finally, usage in the historic literature may vary depending on era and subdiscipline. Therefore, care should be taken when using terms, and we encourage their context definition when used in individual studies. We are strong proponents of including operational definitions for technical terms being used in any study so that readers can clearly understand how the term is applied in specific situations.
Terms and concepts
Absolute error: the difference between the measured or estimated disease severity and the actual value.
Accuracy: closeness of an estimate or measurement to the assumed “gold standard,” real, actual, or true value. Compare to agreement.
Actual value of severity: the most accurately determined value for severity of disease on a specimen against which other measurements, estimates, or predictions are compared (see also true values, “gold standard,” and ground truthing).
Adequate sample size: the number of samples (fields, plants, plant parts, in some cases inoculum of the pathogen) needed to ensure a desired level of accuracy.
Agreement: closeness (or concordance) of two estimates or measurements (same entity, attribute, or outcome and on the same scale) to one another. In cases where one is the assumed actual value, agreement equals accuracy (in other situations where the actual value is not concerned, it is a metric of reliability). Agreement is quantified via different methods depending on the variable type: categorical (nominal or ordinal) or continuous.
Algorithm: a finite sequence of well-defined, computer-implementable instructions that are typically used to solve a class of problems or to perform a computation.
Annotation: the process of adding metadata to a dataset such as when assigning healthy or diseased pixels in a training dataset to the respective class (see training data, see class). This is typically performed manually by human specialists making use of image analysis software.
Artificial intelligence (AI) (as applied to phytopathometry): a computational data-driven approach capable of performing tasks that normally require human intelligence to independently detect, classify, or measure plant disease development, symptoms, and effects on plants. Usually applied in conjunction with the use of imaging sensors. See deep learning and machine learning.
Assessment (disease assessment): the physical process or activity of detecting and quantifying disease — either severity, incidence, or prevalence.
Assessment key: see standard area diagram (the term has been used very rarely to refer to a field assessment key as well).
Assessment protocol: an operational description of how disease intensity in a population will be evaluated.
Assessor: see rater.
Bias: the tendency to overestimate or underestimate disease severity. Bias may be associated with measurement, termed systematic or constant (see systematic bias and constant bias), or with sampling if the sample collected is not representative of the population. In machine learning, data distributions found in the training and test datasets can lead to bias in AI.
Binary: numerical notation of two bases; binary classification typically denotes a simple differentiation of diseased and healthy regardless of the method used during assessment.
Census: when all sampling units within a population (both as operationally defined) are assessed.
Chlorophyll fluorescence: re-emitted signal of incoming light by chlorophyll molecules. Can be measured by imaging and non-imaging fluorescence sensors and provides information on a pathogen’s effect on photosynthesis.
Chlorosis: fading of typical green plant color to light green, yellow, or white due to reduction in chlorophyll density, generally caused by disease or abiotic stresses. Also, see necrosis.
Class (disease class): the numeric value representing a range in disease severity or a progression in symptom severity development (also, see grade; see ordinal scales).
Class (machine learning): a group of sample data with the same semantic (belonging), the classification belongs to supervised machine learning and predicts the class of data points based on training data.
Color transformation: a change in the coordinate system (color space) used to represent colors, usually applied to highlight certain features in a digital image.
Concordance: see agreement.
Confidence interval (CI): a confidence interval for a population sample statistic (e.g., the mean value) that provides an estimate of reliability. The CI represents the range of values for that population sample statistic for which the difference between the statistic and an observed estimate is not significant at the 5% level using a 95% interval corresponding to a P value = 0.05.
Constant bias: the overestimation or underestimation of disease severity is a constant regardless of magnitude of the actual values.
Convolutional neural network (CNN): the most common class of deep neural networks used in deep learning; this type of architecture can be a regularized version of multilayer perceptrons (models devised to represent or simulate the ability of the brain to recognize and discriminate), thus being more robust to overfitting.
Correlation: the presence of an association between two different measures or variables. Correlation is not synonymous with agreement. Two sets of observations, which are highly correlated, may have poor agreement; however, if the two sets of values agree, they will surely be highly correlated. Compare to the concordance correlation coefficient.
Count (disease count or lesion count): the number of lesions per plant, plant organ, or unit area (one of the ways in which plant disease severity may be quantified).
Cross-validation: a technique often used to assess the generalization capability of a model. In k-fold cross-validation, a given dataset is divided into k subsets, from which one is retained for testing and the remainder k-1 subsets are used for training. The process is repeated k times (folds), always using a different subset for testing.
Data fusion: the process in which different types of data are combined in order to provide results that could not be achieved using single data sources (e.g., combining images and meteorological data to provide accurate detection of a specific disease).
Deep learning: a special case of the machine learning model that utilizes artificial neural networks that abstract basic parameters about the data and subsequently train the computer to learn by recognizing patterns using many layers of processing, in this case, to detect or measure plant disease. Deep learning is appropriate for large datasets with complex features and where there are unknown relationships within the data (see artificial intelligence, machine learning, supervised learning, and unsupervised learning).
Density (disease density, lesion density): a measure of disease severity. Disease counts may be referred to as disease density because the counts are expressed relative to an organ; incidence, percent severity, and counts may all be expressed as densities based on quantity of disease per unit area (e.g., per unit area of crop).
Destructive sampling: sampling whereby the sampling unit is removed from the population, with the sampling unit being used to obtain disease or pathogen measurements.
Detection (disease detection): determination that disease is present on a specimen without quantification of disease intensity. May be visual or sensor based (or by molecular means).
Detection accuracy: in sensor-based systems, detection accuracy is defined as \( \frac{true\ positive+ true\ negative}{total\ sample\ (n)} \). It is important to note that the accuracy defined here is different from accuracy as described in measurement science or as defined in statistics.
Detection assay: an assay to determine whether some biological entity of interest is present in a sampling unit.
Detection threshold: the minimum quantity of disease or pathogen intensity (prevalence, incidence, severity) needed for a rater, machine, or protocol to accurately detect the presence of a disease or pathogen.
Diagrammatic key: see standard area diagram.
Diagrammatic scale: see standard area diagram.
Disease app: an interactive device-based application using image analysis to aid disease assessment and that may also measure the symptomatic area caused as a result of infection by a plant pathogen.
Disease diagram: a pictorial or graphical representation, usually a drawing, of a specimen with symptoms where the relative area affected is unknown. Compare to the standard area diagram.
Disease severity index: a disease severity index (DSI) is calculated based on the frequency of estimates of classes on an ordinal scale in the sampling units. There are variations on the theme, but often DSIs are calculated as DSI = (Σ(class frequency × score of rating class))/((total number of observations) × (maximal disease index)) × 100. Also, see ordinal scales and infection index.
Disease: the abnormal functioning of an organism, often manifested by visible symptoms which can be quantified. The cause may be biotic (caused by a pathogen) or abiotic (caused by a nutrient deficiency or toxicity). In phytopathometry, the cause of disease is biotic due to pathogenic microorganisms.
Distribution map: spatial distribution of a disease within a crop stand or field (see mapping).
Drone: See unmanned aerial vehicle (UAV).
Early detection: assessment of diseases by sensors (or molecular methods) before symptoms are visible to the naked eye.
Estimate (or estimation): a visual raters’ quantitative (numeric) approximation of the presumed actual disease severity (measured using a standard method, for example, red–green–blue (RGB) image analysis) on a specimen (compare to measurement).
Feature: measurement of a specific property of a data sample. It can be a color, texture, shape, reflectance intensity, index values, or spatial information.
Field assessment key (or field key): a rating scale to facilitate the rapid assessment of disease in defined crop plantings (plots or fields) rather than on individual plants or plant organs. Quantities may be based on numeric or qualitative descriptors or a combination based on the perception of disease at that spatial scale.
Forensic sampling: the act, process, or technique of selecting an appropriate sample relating to the use of science or technology in the investigation and establishment of facts or evidence in a court of law.
Geometric three-dimensional (3D) sensor: 3D sensors measure the geometric surface of objects spatially, resulting in a 3D point cloud. This can be used to extract size and growth parameters of plants and plant organs. See light detection and ranging (LiDAR).
Geometry: shape properties of a specimen expressed as position and relation of points, lines, and surfaces in a tridimensional space. Object geometry has a marked effect on disease assessment by sensors, emphasizing the need for imaging setups to include sensors for geometry and reflectance. Geometry can be measured using images in two dimensions (shape) or by using three-dimensional measuring devices.
Gold standard: see actual value.
Grade: see class.
Ground truthing: the term used in most sensor-based studies where the sensor-based measurements are compared to the assumed actual values (see actual value) or reference data.
Growth stage key: a descriptive or pictorial representation of specific crop growth (or developmental) stages, commonly used as a phenologic reference to when disease assessments were conducted.
High-throughput screening: systematic, automated, and noninvasive assessment of plant traits or diseases for a large number of genotypes.
Hue, saturation, and intensity (HSI) color model: represents every color in red–green–blue (RGB) color space with three components, namely, hue, saturation, and intensity. Hue describes the color in the form of an angle (0 to 360°); saturation describes how much the color is diluted with white light (0 to 1), and intensity ranges from white to black (0 to 1). The concept is used extensively in visible spectrum image analysis.
Hyperspectral imaging: the process of using a spectral imaging sensor to collect and process reflectance information from the electromagnetic spectrum to obtain a spectral signature of each pixel in the image of the specimen. Hyperspectral imaging typically assesses several hundred narrow wavebands extending beyond the visible spectrum. The resulting spectral signatures of healthy and diseased plant pixels vary in a specific way.
Identification (disease identification): the correct assignment of the cause of symptoms of the plant disease (the disease name, and causal pathogen or condition). Compare to detection.
Image analysis: the detection, identification, and quantification of disease on specimens by extracting the appropriate feature (see feature) or information from digital images using a variety of image processing tools and techniques that allow delineation and separation of diseased and healthy areas. Pixel counts are most often converted to percentage areas.
Image preprocessing: the activities to enhance/prepare the quality of digital images of specimens for further analysis. This may include converting an image to binary or grayscale, calibration, undistortion, or removing speculations.
Incidence (disease incidence): the proportion of specimens diseased (regardless of the method) of the total number of specimens assessed (in other branches of science including medicine and entomology, this equates to prevalence. See definition of prevalence as used in plant pathology).
Infection index: see disease severity index.
Injury: damage caused to a plant or plant part by an agent including herbivores, pathogens, chemicals, or unfavorable environmental conditions.
Intensity (disease intensity): a general and broad term to encompass the quantity of disease expressed as incidence, prevalence, area diseased, density, or counts.
Inter-rater reliability: a statistical measure of consistency of disease assessments across the same specimens between raters. See also reproducibility.
Intra-rater reliability: a statistical measure of consistency of disease estimates on the same specimens by the same rater or measures of severity on the same specimens by the same sensor or instrument. See also repeatability.
Just noticeable difference: also known as the difference threshold. In psychometrics, the just noticeable difference (JND) is the minimum level of stimulation that an individual can visually detect 50% of the time. In phytopathometry, the JND translates to the minimum difference in severity that can be visually perceived between two specimens.
Lesion: a localized externally visible diseased area or wound.
Light detection and ranging (LiDAR): in ground-based LiDAR, a pulsed laser is sent to an object, and when combined, they generate accurate 3D information regarding the target object. LiDAR has been utilized as a tool in some plant disease detection studies. LiDAR is also known as terrestrial laser scanning (TLS).
Location shift: see constant bias.
Machine learning: application of artificial intelligence (AI) algorithms that underpin the ability to learn characteristics of a disease (or lesion) via extraction of features (see feature) from a large dataset. Machine learning models are often based on knowledge obtained from annotated training data (specimen images previously assigned to a distinct class). Once the model is developed, it can be used to predict severity (specimen or pixel value) on test data or unknown images (see artificial intelligence and deep learning).
Mapping: a combination of information from phytopathometry with geographic information, indicating the spatial distribution of a disease.
Mathematical morphology: this type of technique, usually applied to binary images, can simplify image data, eliminating irrelevant structures while retaining the basic shapes of the objects. All morphological operations are based on two basic operators, dilation and erosion (dilation is a process of adding pixels to the boundaries of objects in an image, while erosion is the process of removing pixels on object edges).
Measurement: the area, size, length, or amount of a variable (in this case disease on a specimen), as established by measuring the characteristics or severity using a device (nonhuman) or sensor. If a rater were to use a ruler to measure a symptom, it would be considered a measurement (compare to estimate).
Multispectral imaging: the sensor-based collections and processing of reflectance information from several wavebands of the electromagnetic spectrum. Typically, up to 10 wavebands in the visible range or near infrared range are assessed as data for disease detection.
Multi-temporal assessment: repeated, noninvasive (nondestructive) assessment of a specimen/object over a specific time period (daily, weekly, monthly) to monitor disease progress or disease dynamics.
Necrosis: the death of cells or tissue that results in darkening of the tissue to black or brown that may be typical of some disease symptoms. Also, see chlorosis.
Nominal scale: a measure of the quantity of disease based solely on a qualitative label (e.g., no disease “−,” a little disease “+,” moderate disease “++,” severe disease “+++”).
Nondestructive sampling: a method of collecting information whereby the sampling unit is not removed from the population, also called noninvasive sampling.
Ordinal scale: the quantity of disease based on a rank-ordered series of values (e.g., 0, 1, 2, 3, 4, where 4 is the most diseased state), defined qualitatively or quantitatively.
Pathometry (plant pathometry): see phytopathometry.
Phytopathometry: the branch of plant pathology, traditionally studied as part of plant disease epidemiology, tasked with the science of plant disease measurement.
Plant phenotyping: the quantitative assessment of plant traits, in this case disease intensity. Recently, digital phenotyping is a synonym for the use of sensors, robotics, machine learning, and artificial intelligence. Plant phenotyping links plant pathology with plant breeding in the genomics era.
Precision: statistically a measure of the variability of estimates or measurements of disease on the same specimens obtained by different raters or instruments. However, in measurement science, precision is not just a measure of variability when considering reliability or accuracy. Reliability and accuracy also account for the concordance between the estimates and measurements as well as the variability.
Prevalence (disease prevalence): the proportion or number of fields or geographic units (municipalities, counties, etc.) where the disease has been detected (on at least one specimen).
Proximal sensing: optical information of a crop specimen obtained under controlled conditions, without physical contact with the specimen but at relatively close distances, often in the greenhouse or laboratory. Scales considered are plant, leaf, or cell; a disease or pathogen signs (spores, mycelia, etc.) may also be detected. Proximal sensing may be noninvasive or semi-invasive.
Psychophysics: the branch of psychology that deals with the relationships between physical stimuli and mental phenomena, in this case the perception of plant disease and estimation of the quantity, usually in relative terms (percent area affected), present on a specimen.
Qualitative ordinal scale: a scale where each class in the ranked order is defined by a qualitative description of progressively more advanced symptoms of disease.
Quantitative ordinal scale: a scale where each class in the ranked order is defined by a numeric, increasing sequential range (usually on the percentage scale) indicating more intense disease symptoms based on the relative area diseased.
Rater: an individual who assesses disease. See also assessor.
Rating scale: description in words or numbers of disease classes used in scoring plant parts, plants, plots, or fields (usually ranging from no disease to completely diseased).
Rating: the estimate or measurement of disease severity on a particular specimen. An appropriate disease class, value, or code number assigned to each sampling unit in a sample population to indicate the quantity of disease.
Ratio scale: a numeric scale used to rate severity of disease on a specimen and is characterized by equal-sized intervals and a defined zero. The percentage scale is an example of a ratio scale.
Red–green–blue (RGB) color space: additive color model in which red, green, and blue are added together to reproduce a wide range of colors. This representation is used in most visible spectrum sensors.
Reflectance: the proportion of the incident light in the electromagnetic spectrum reflected by an object per wave band.
Relative error: the absolute error (see definition) divided by the actual measurement. May be presented as a percentage (where it is called the percentage error).
Reliability: the extent to which the same estimates or measurements of diseased specimens obtained under different conditions yield similar results. See also inter-rater reliability, intra-rater reliability, repeatability, and reproducibility.
Remote sensing: optical information of an object obtained in the field or landscape, in a noninvasive manner, without contact with the sampling units using a sensor (which could be the human eye or an artificial spectral sensor).
Repeatability: see intra-rater reliability.
Reproducibility: see inter-rater reliability.
Resistance class: description of susceptibility based on intensity of development of disease symptoms in different genotypes or under different environments in a plant species. Terms are typically “susceptible,” “moderately susceptible,” “moderately resistant,” and “resistant,” or variations thereon. May be derived from a scale where particular classes are grouped accordingly or applied directly.
Sample unit: the individual (e.g., a leaf or whole plant) member of a defined population (e.g., plant, field, region) that is assessed for disease. In all cases, it must be defined in any study. A certain number of sample units will comprise the sample size, which must also be defined.
Sample: in plant disease assessment, a sample is a set of units (e.g., leaves, fruit, plants) collected or selected from a population by a defined procedure.
Sampling protocol: an operational description of how sampling units will be selected from a population.
Scale shift: see systematic bias.
Scale: one of a number of methods to quantify disease (nominal, ordinal, and ratio scales). May also be the physical level of assessment from cell to leaf to plant to canopy and distance from object to sensor (see remote sensing/proximal sensing).
Scoring: see assessment.
Segmentation: the process of partitioning a digital image of a diseased specimen into multiple segments of similarity or classes (based on sets of pixels with common characteristics of hue, saturation, and intensity) either automatically or manually. In the latter case, the human-powered task is also called image annotation in the context of training AI algorithms. For example, the process may include the background, leaf specimen, and diseased area on the leaf specimen. When performed at the pixel level, the process is referred to as semantic segmentation. Also, see thresholding and classes.
Sensitivity: also called the true positive rate, measures the proportion of positives (e.g., diseased) that are correctly identified. Used as a statistical measure of the performance of a binary classification test. Also known as recall or probability of detection in machine learning. Compare to specificity.
Sensor data fusion: combination of data from different sensor types to accurately assess diseases on plants (e.g., geometric and reflectance sensors).
Severity (disease severity): degree to which a specimen (plant, plant part, or plot) is diseased. Severity may be described quantitatively on a scale (for example, lesion length in mm, or lesion area in mm2), or as a proportion of the unit area diseased (commonly a percentage), or as the number of lesions present, or as a ranked numeric order (i) based on intervals of the percentage scale, or (ii) based on descriptions of symptoms in a progression from mild to severe (as applicable for many systemic diseases). This definition differs from most prior definitions that described severity purely on the basis of relative area diseased — many plant diseases show degrees of severity without an easily defined area affected (e.g., many systemic diseases), hence the revised broader definition.
Sign: an indication of disease from direct observation of a pathogen, its parts (e.g., mycelia of powdery mildews on the leaf surface), or sensorial abnormality (e.g., softness, smell). Also, see symptom.
Specificity: also called the true negative rate, measures the proportion of negatives (e.g., non-diseased) that are correctly identified as such. Used as a statistical measure of the performance of a binary classification test, commonly used in remote sensing. Compare to sensitivity.
Specimen: a unit of assessment in a sample (usually a leaf, fruit, stem, root, flower or plant, or other plant part). Exceptionally, a plot or field. Also, see sample unit.
Spectral signature: reflectance intensities over the electromagnetic spectrum which are characteristic for a specific disease or developing stage of disease.
Standard area diagram set: a generic term for a pictorial or graphic representation (drawing or true-color photo) of selected disease severities on plants or plant parts (leaves, fruit, flowers, etc.) generally used as an aid for more accurate visual estimation (on the percentage scale) or classification (using an ordinal scale) of severity on a specimen. Standard area diagram sets, commonly abbreviated to SADs, are usually presented as a set of diagrams for interpolation using the percentage scale. See diagrammatic scale and disease diagram.
Supervised learning: a machine learning model, based on a known labeled training dataset that is able to predict a class label (classification) or numeric value (regression) for new unknown data (see also unsupervised learning).
Survey: a process of determining the prevalence, incidence, or severity of a disease in an operationally defined population with sampling units selected to represent the spatial scale of interest.
Symptom: an indication of disease by the reaction of the host, e.g., canker, leaf spot, scab, and wilt (also, see the definition of “sign” for other diseases that may manifest through pathogen structures without inducing symptoms in the host).
Systematic bias: the overestimation or underestimation of disease severity is dependent on the magnitude of the actual value.
Thresholding: the simplest method of segmenting an image and analyzing sensor data. The process of creating a binary differentiation of data or an image based on selecting a value on the pixel intensity of the original specimen image to separate the healthy and diseased tissue. Also, see segmentation.
Trait: genetically determined characteristic that can influence symptom expression in a characteristic way.
True value: see actual value.
Unmanned aerial vehicle (UAV): also known as an unmanned aerial system (UAS) or drone is any aircraft that can be controlled without a human pilot on board. The UAV is a platform for optical or similar sensors with imaging capabilities. UAVs are being increasingly explored in the context of phytopathometry.
Unsupervised learning: structure and similarities of data are clustered to reveal group affiliations in an unknown and new dataset. The ground truth or the true group annotation is unknown during clustering (see also supervised learning).
Virtual lesion: a fraction of the leaf, usually green and surrounding a visible lesion, where photosynthesis is null.
Visible (VIS) spectrum: it is the portion of the electromagnetic spectrum that is visible to the human eye, corresponding to wavelengths between 380 and 750 nm.
Visual assessment: the process of estimating disease severity and incidence based on eyesight, perception, classification, and approximation of disease severity on a plant specimen(s).
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Agrios GN (2005) Plant pathology, 5th edn. Elsevier Academic Press, Burlington, MA
Behmann J, Mahlein A-K, Rumpf T, Römer C, Plümer L (2015) A review of advanced machine learning methods for the detection of biotic stress in precision crop protection. Precision Agriculture 16:239–260
Bock CH, Poole GH, Parker PE, Gottwald TR (2010) Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Critical Reviews in Plant Sciences 29:59–107
Bock CH, Chiang K-S, Del Ponte EM (2016) Accuracy of plant specimen disease severity estimates: concepts, history, methods, ramifications and challenges for the future. CABI Reviews 11:1–21
Bock CH, Hotchkiss MW, Brenneman TB, Stevenson KL, Goff WD, Smith MW, Wells L, Wood BW (2017) Severity of scab and its effects on fruit weight in mechanically hedge-pruned and topped pecan trees. Plant Disease 101:785–793
Bock CH, Barbedo JGA, Del Ponte EM, Bohnenkamp D, Mahlein A-K (2020) From visual estimates to fully automated sensor-based measurements of plant disease severity: status and challenges for improving accuracy. Phytopathology Research 2:1–36
Chiang K-S, Liu HI, Chen YL, El Jarroudi M, Bock CH (2020) Quantitative ordinal scale estimates of plant disease severity: comparing treatments using a proportional odds model. Phytopathology 110:734–743
Cook G, Coetzee B, Bester R, Breytenbach JHJ, Steyn C, de Bruyn R, Burger JT, Maree HJ (2020) Citrus tristeza virus isolates of the same genotype differ in stem pitting severity in grapefruit. Plant Disease 104:2362–2368
Cooke BM, Gareth-Jones D, Kaye B (2006) The epidemiology of plant diseases, 2nd edn. Springer, Dordrecht, The Netherlands
Colson ES, Platz GJ, Usher TR (2003) Fungicidal control of Pyrenophora tritici-repentis in wheat. Australas Plant Pathol 32:241–246
Cowling WA, Gilchrist DG (1980) Influence of the pathogen on disease severity in Stemphylium leafspot of alfalfa in California. Phytopathology 70:1148–1153
D'Arcy CJ, Eastburn DM, Schumann GL (2001) Illustrated glossary of plant pathology. The Plant Health Instructor. https://www.apsnet.org/edcenter/resources/illglossary/Pages/default.aspx. Accessed 25 Jun 2021
Del Ponte EM, Pethybridge SJ, Bock CH, Michereff SJ, Machado FJ, Spolti P (2017) Standard area diagrams for aiding severity estimation: scientometrics, pathosystems, and methodological trends in the last 25 years. Phytopathology 107:1161–1174
Everitt BS (1998) The Cambridge dictionary of statistics. Cambridge University Press, Cambridge, UK, p 360
Gottwald TR, Aubert B, Xue-Yuan Z (1989) Preliminary analysis of citrus greening (Huanglungbin) epidemics in the People’s Republic of China and French Reunion Island. Phytopathology 79:687–693
Gregory PH (1982) E. C. Large, and phytopathometry. Plant Pathology 31:7–8
Heim RHJ, Carnegie AJ, Pablo J, Zarco-Tejada P (2019) Breaking down barriers between remote sensing and plant pathology. Tropical Plant Pathology 44:398–400
Horsfall JG, Barratt RW (1945) An improved grading system for measuring plant disease. (Abstr.) Phytopathology 35:655
Kousik CS, Mandal M, Hassell R (2018) Powdery mildew resistant rootstocks that impart tolerance to grafted susceptible watermelon scion seedlings. Plant Disease 102:1290–1298
Large EC (1953) Some recent developments in fungus disease survey work in England and Wales. The Annals of Applied Biology 40:594–599
Large EC (1966) Measuring plant disease. Annual Review of Phytopathology 4:9–26
Ling K-S, Scott JW (2007) Sources of resistance to Pepino mosaic virus in tomato accessions. Plant Disease 91:749–753
Lucas JA (2002) Plant pathology and plant pathogens, 4th edn. John Wiley & Sons, Ltd., Chichester, West Sussex, UK
Madden LV, Hughes G, van den Bosch F (2007) The study of plant disease epidemics. APS Press, St. Paul, MN, USA
Mahlein A-K (2016) Plant disease detection by imaging sensors - parallels and specific demands for precision agriculture and plant phenotyping. Plant Disease 100:241–251
McRoberts N, Hughes G, Madden LV (2003) The theoretical basis and practical application of relationships between different disease intensity measurements in plants. The Annals of Applied Biology 142:191–211
Nutter FW Jr (2001) Disease assessment. In: Maloy OC, Murray TD (eds) Encyclopedia of plant pathology. John Wiley and Sons, Inc., New York, NY, pp 312–323
Nutter FW Jr, Teng PS, Shokes FM (1991) Disease assessment terms and concepts. Plant Disease 75:1187–1188
Nutter FW Jr, Esker PD, Coelho Netto RA (2006) Disease assessment concepts in plant pathology. European Journal of Plant Pathology 115:95–103
Pascual A, Campa A, Pérez-Vega E, Giraldez R, Miklas PN, Ferreira JJ (2010) Screening common bean for resistance to four Sclerotinia sclerotiorum isolates collected in northern Spain. Plant Disease 94:885–890
Paulus S (2019) Measuring crops in 3D: using geometry for plant phenotyping. Plant Methods 15:103
Paulus S, Mahlein A-K (2020) Technical workflows for hyperspectral plant image assessment and processing on the greenhouse and laboratory scale. Gigascience 9(8): giaa090. https://doi.org/10.1093/gigascience/giaa090
Pethybridge SJ, Sharma S, Hansen Z, Kikkert JR, Olmstead DL, Hanson LE (2020) Optimizing Cercospora leaf spot control in table beet using action thresholds and disease forecasting. Plant Disease 104:1831–1840
Scherm H, Christiano R, Esker PD, Del Ponte EM, Godoy CV (2009) Quantitative review of fungicide efficacy trials for managing soybean rust in Brazil. Crop Protection 28:774–782
Seem RC (1984) Disease incidence and severity relationships. Annu Rev Phytopathol 22:133–50
Tronsmo AM, Collinge DB, Djurle A, Munk L, Yuen J, Tronsmo A (2020) Plant pathology and plant diseases. CABI, Wallingford, UK
Urrea CA, Harveson RM (2014) Identification of sources of bacterial wilt resistance in common bean (Phaseolus vulgaris). Plant Disease 98:973–976
Wang D, Wang B, Wang J, Wang S, Wang W, Niu Y (2020) Exogenous application of harpin protein Hpa1 onto Pinellia ternata induces systemic resistance against tobacco mosaic virus. Phytopathology 110:1189–1198
Funding
Clive H. Bock was funded by the USDA-ARS National Programs through CRIS project 6042-21220-014-00. Sarah J. Pethybridge was supported by the United States Department of Agriculture National Institute of Food and Agriculture (USDA-NIFA) Hatch project NYG-625424. Jayme G. A. Barbedo was supported by the Brazilian Agricultural Research Corporation — Embrapa (SEG 02.14.09.001.00.00) and by the São Paulo Research Foundation — FAPESP (proc. 2013/06884-8). Paul D. Esker was funded by the USDA National Institute of Food and Federal Appropriations under Project PEN04660 and Accession Number 1016474. Anne-Katrin Mahlein was partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2070-390732324 and the Farmerspace project which is supported by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany. The Federal Office for Agriculture and Food (BLE) provides coordinating support for digitalization in agriculture as a funding organization, grant number FZK 28DE104A18. Emerson M. Del Ponte was supported by the National Council for Scientific and Technological Development (CNPq) through a Productivity Research Fellowship (PQ) project 310208/2019-0.
Author information
Authors and Affiliations
Contributions
Conceptualization: CHB and EMDP. Methodology: CHB and EMDP. Resources: CHB, SJP, JGAB, PDE, AKM, EMDP. Writing — original draft preparation: CHB and EMDP. Writing — review and editing: CHB, SJP, JGAB, PDE, AKM, and EMDP. Visualization: CHB and EMDP. Project administration: CHB. Funding acquisition: CHB, SJP, JGAB, PDE, AKM, and EMDP. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bock, C.H., Pethybridge, S.J., Barbedo, J.G.A. et al. A phytopathometry glossary for the twenty-first century: towards consistency and precision in intra- and inter-disciplinary dialogues. Trop. plant pathol. 47, 14–24 (2022). https://doi.org/10.1007/s40858-021-00454-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-021-00454-0