Abstract
Purpose
B-mode ultrasound images are used in identifying the presence of fat deposit if any in carotid artery. The intima media, lumen, bifurcation boundary is detected by the echogenic characteristics embedded in the carotid artery.
Methods
A fully automatic self-learning based segmentation is proposed by extracting the edges by a modified affinity propagation, which are given as inputs to the Density Based Spatial Clustering of Applications with Noise (DBSCAN) for super pixel segmentation. The segmented results are analyzed with Gradient Vector Flow (GVF) snake model and Particle Swarm Optimization (PSO) clustering based segmentation using various performance measures.
Results
The proposed parameter free, fully automatic segmentation method combining Affinity propagation and DBSCAN are evaluated for a database of 361 images and gives reinforced results in the longitudinal B-mode ultrasound images. The proposed approach gives an improved accuracy of 12% increase when compared with the manual segmentation and 15% compared with segmentation by affinity propagation and DBSCAN when performed individually. The average Root Mean Square Error (RMSE) is 110 ± 44 µm.
Conclusion
Extracted edge points are used for clustering in a fully automated carotid artery segmentation approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Ultrasound imaging is a promising imaging modality in understanding the characteristics of carotid arteries and related vascular problems. The deposit of fats in the artery walls results in atherosclerosis and other cardio-vascular diseases like stroke, myocardial infarction and heart attack. B-mode ultrasound images have benefits of low signal to noise ratio (SNR) and spatial resolution compared to Magnetic Resonance imaging (MRI) and Computer Tomography (CT). Avoidance of radiation exposure and injection of contrast agents, highly economical, portable, real time are the added advantages of ultrasound imaging. B-mode imaging is used in practice since it gives anatomical and structural details of the artery.
Manual Intima- Media Thickness (IMT) measurement by skilful sonographers is not consistent over time and is time consuming. The outcomes depends on the individual and is subjective. Semi-automatic measurements were proposed, which requires the sonographer to select the seed points for segmentation [1, 2]. Automation provides more advantages than manual and semi-automatic techniques for huge population. After 2007, user independent systems for identification of plaque started to evolve, which led to faster segmentations [3, 4]. There were nearly 15% failure probabilities and was not suitable for real time situations. Various protocols and standardizations were proposed for computer aided systems to help operators in measuring IMT. Some of them were motion estimation, stiffness measurement, blood flow analysis, 2D, 3D, doppler image analysis [5,6,7].
Contour detection based on edgelet information with a Bayesian modelling is proposed [8, 9]. Soft delineation recognition map is derived from the estimated trajectory delivery using a particle filter. The method requires manual adjustments and contour framing. Level set methods were introduced, which moves the initial contours towards the artery wall by reducing an energy function depending on edge detection. The gradient value and the difference between the images were used as metrics for measurements. Accurate detection of missing contour points with exact shape and size constraints is performed, which can avoid discontinuities in boundaries [10]. In this approach, the vessel boundary initialization is determined by a radius function, assuming a circular cross-sectional plane for vessel, which may not be suitable in all conditions. This non-linear least square problem can be solved by self-learning techniques for segmentation [11].
Andres et al. proposed a 3D coupled ideal graph cut procedure identifies the borders into a graph cut using the information from multiple sequences [12]. Three seed points were selected in the external, internal and common carotid artery. The dissimilarity among the left and the right sides can be identified after a snake segmentation of the CCA. This paper proves that measurement of the IMT thickness in any one side is sufficient for normal people, since both are symmetrical. With increasing age, IM thickness measure increases. Patients with Cardio-Vascular Diseases (CVD) shows significant statistical differences in the right and left IMT measurements [7].
The texture feature varies considerably between the cardiac systolic and diastolic stages in the ultrasound video of the carotid plaque [13]. Combining the systolic and diastolic features gives improved results than using them alone. Modified semi-supervised affinity propagation approach with autonomous module study were developed for clustering the ECG heart beat recordings [14]. By this method, the labelled samples were used as seeds to initialize the cluster centers in k-means algorithm. The method does not use random sampling and so it is well suited for real time situations where incorrect initializations and hard decisions may impact the solution.
Jing et al. proposed a illustrative multi-label learning approach was developed to reject the effect of noisy features and select only representative features [15]. It was applied in affinity propagation with Support Vector Machine (SVM) to frame the relationship among the designated features and for the learning process. The non-metric similarity between the pictures by soft matching of Scale Invariant Feature Transform (SIFT) was proposed, which gave significant results than by hard matching, which used binary representation [16]. SIFT can recognize look features, which are illumination, 3D camera view point invariant, scale and rotation. Chen et al. proposed an approach where individual features class discrimination ability can be extended within a new sub-space, minimizing the spectral correlation among the chunklets features, followed by affinity propagation based clustering [17]. By learning a discriminative transformation, a standardized metric function is assimilated, using the positive and negative constraints. Clustering exemplars are obtained preserving low correlation and high separable data points. Ground truth label information for data points are not required, making it suitable for any image.
A small number of labelled exemplars are incorporated into the affinity propagation in a semi-supervised technique [18]. Similarity matrix is adjusted by the labelled samples and the unlabeled data selection and needless label reduction are done using incremental and decremental affinity propagation technique. Thus learning bias and stability-plasticity dilemma are evaded with a drawback of increased computational complexity.
Based on a Fuzzy Statistical Similarity (FSS) quantity, affinity propagation based clustering was proposed which shows faster implementation, least error and demonstrates how near two pixel vectors look alike [19]. In this method, the data points are taken as applicant exemplars and permits soft data till a subset of data points become exemplars. Incorrect initialization and hard decision problems reduces the computational complexity and gives ideal results.
DBSCAN algorithm with geometric restrictions and color similarity clusters the pixels and merges clusters into small super-pixels by their neighboring pixels through a distance measure using spatial and color features [20]. Centroid of the cluster identification was done using the number of pixels and cumulative values, which are used to build Color Sum of Absolute Differences (CSAD) to identify the decision criteria and the difference measure [6]. The method was much suitable for images which has color information for better area matching.
Jian et al. proposed a parameter less algorithm was proposed depending on dominant sets and DBSCAN. User independent parameters Minpts are identified with respect to clusters from DSets in arbitrary shapes [21]. The method solves the over-segmentation effect, with number of clusters closer to ground truth value, increasing the cluster size. A modified DBSCAN is used to create saliency map with the sensed traffic light followed by template matching [22].
After setting an initial boundary, three cluster centers are approximately computed using level set technique [7]. From the resulting cluster centers, level set density map is created and final segmentation is done with valley seeking clustering. During clustering, outlier detection is also performed at the same time using the initial boundary set during the level set method. The method avoids the over-fitting problem and also correctly classifies the cluster centers and outliers.
To cope up with this, a parameter free segmentation approach where the edge points of the image are extracted using affinity propagation is modelled. With extracted edges as core and border points, a local cluster based segmentation with DBSCAN is proposed. Both the algorithms are made parameter free thus making it not relying on initial assumptions and ground truth. The methods prevents interior area shrinkage and can be combined with any fitting terms.
The paper layout is as follows. Carotid artery ultrasound imaging and its significance in IMT measurement is explained in Sect. 2. Affinity propagation based edge detection in Sect. 3 followed by DBSCAN based clustering in Sect. 4. Section 5 explains PSO and GVF based segmentation, Sect. 6 gives the experimental dataset, performance measures, Sect. 7 portrays the results and discussions and Sect. 8 concludes the paper.
2 Carotid Artery Ultrasound Imaging
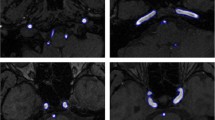
Standard B-mode Ultrasound Imaging (BMUS) is used for lumen segmentation to identify abnormalities in carotid artery. Vulnerable plaques possess some different biological, mechanical and morphological structures, which has to be identified to prevent further rupture. Uneven lumen shape, noise in lumen and echolucent plaque are some problems faced during the segmentation process. Figure 1 shows the example ultrasound images of the carotid artery with and without plaque.
BMUS segmentation were done using methods like deformable active contours, Bayesian model, Transforms, and other classification based approaches [5]. Studies proves that primary artery stiffness causes vascular variations and predicts major vascular diseases [4, 23]. Ultrasound imaging uses mechanical energy and is repeatable, highly economic, reliable and extremely safe for patients. Carotid intima- media, lumen thickness is necessary for early detection of abnormalities [3]. A suitable Region of Interest selection is necessary for starting the segmentation process [24]. Factors like age, gender, hemodynamics, blood glucose level, lipid level take diverse properties in the left and right CCA [25, 26].
Completely automated carotid artery segmentation gives better accuracy in identifying the random shaper lumen, good repeatability, better efficiency, reduced computational cost, tractability and error control [27]. Though carotid wall irregularity can identify atherosclerosis, IMT measurement gives exactly the extent of the deposits in the artery and the risk factors [28].
3 Affinity Propagation Based Segmentation
When poorly visualized, the speckle noise in the ultrasound image makes it tough to identify the lumen boundary. Lipid rich plaque formation or a transducer not angled properly may further affect the image quality. Speckle noise is the inherent artifact in ultrasound image and is denoised using curvelet filtering, which is a multi-scale analyzer. Taking edge as an indicative, the objective function being the characteristics of a singular curve, line singularity is mapped as point singularity [29].
Bigger curvelet constants will have additional coefficients at its neighborhood positions. The curvelet transform has acceptable scale directional components and rough scale isotropic father wavelet [30]. Curvelet soft thresholding gives best denoised results and also reserves maximum of the necessary edge features. By this method, both the smooth low frequency noise and the oscillatory high frequency noise are filtered and given as input to the segmentation algorithm. Figure 2 gives the proposed method for the affinity propagation and DBSCAN based segmentation. The extracted noise coefficients, subjected to curvelet soft thresholding results in the denoised coefficients in the approximate, coarse and fine scales. Inverse curvelet transform is to bring back the image from the radon domain to the transform domain. The denoised image is given to affinity propagation for edge detection and is to be fed as input to DBSCAN for clustering based segmentation. Figure 3 gives the sample carotid artery ultrasound image and denoised with curvelet decomposition.
Affinity propagation clustering depends on message passing between data points, which do not need identifying the number of clusters prior. While passing message within a pair of data points, exemplars arises and each one is identified as a cluster. Similarity among the data points for how it is appropriate to be an exemplar for other data points is assigned as input. If two data points are not similar, it may be omitted or assigned as infinity. Preferences are assumed, on how well each data points are appropriate to the exemplar. Any information which may be required while clustering can be included in the preferences. With applications having large number of clusters, the method is fast, generally applicable -and has better performance [14, 31]. Similarity (S), Responsibility (R) and Availability (A) matrices are updated iteratively to extract the final criteria value. Figure 4 gives the flow diagram of affinity propagation algorithm.
Affinity propagation algorithm requires quadratic memory but has minimum error, faster speed and is flexible. Initially the data point sets are transformed into a distance matrix and the point densities are identified rendering to the adjacent neighbour distance matrix.
In the algorithm given below, α(Ck) is the sum over all the configurations sustaining fi given ck. The first two largest memory elements and the cumulative sum are identified to convert the sum_product affinity propagation to max_ products, in which the numerical precision inaccuracies are removed. Availabilities are initially set to zero so that the difference between the responsibility r(i,k) is made equal to the input similarity among points i and k subtracted from the biggest similarity among i and eligible exemplars. Figure 5 gives the core, border and noise points in the image.

How many other points belong to that exemplar is known after more number of iterations and their availability status becomes less than zero. Thus their cumulative value reduces and it will be slowly removed from the potential exemplars list. Self—availability a(j,j) is rationalized by summing up the positive values of the new responsibilities leaving the threshold value. A lower value for preference gives few number of clusters and a higher value increases the number of clusters. The criteria matrix is represented as
r and a are responsibility and availability values at points (u,v).
The dividing point of data points is first identified and the edge detection is initiated from that point. Initially, a rational partition is obtained where the data point in a cluster is very distinct from data points of other clusters. The time complexity is order of (N × N log N), considering each pixel as potential exemplar. For a 512 × 512 image, the count of data points is 262144 and the complexity becomes 512 × 512 × log 512. To reduce the computational complexity the number of data points has to be minimized without losing the potential exemplars.
4 DBSCAN Based Clustering
Density based spatial clustering of applications with noise (DBSCAN) is a non-parametric density dependent clustering approach where it clusters spatially close points based on their density similarity. A low density point in the neighbourhood may be marked as an outlier provided its similar density points are farther away. Figure 6 gives the flow diagram of DBSCAN algorithm.
Points Marking process in DBSCAN algorithm. The histogram equalized image of the pair wise equivalent data make it independent of initial initializations. As the data points from affinity propagation is an added input, the proposed approach becomes parameter free. a Standard DBSCAN search. b Proposed approach combining affinity propagation edge points and DBSCAN clustering
The points are defined as follows. A point can be considered a core point if the minpoints are within the distance ɛ. If a point q is inside a distance ɛ, it can be considered a directly reachable point from p. In a path p1 to pn, all being core points, with p1 = p and pn = q, each pi+1 accessible from pi, then q point can be reachable from p. The other not reachable points are called outliers or noise points. Two points can be considered as density associated if there is additional point from which the two points are reachable. Thus a cluster contains all the density reachable points alone.
DBSCAN is robust and can cluster the image without initial initialization of the number of clusters. The required parameters minpts and ɛ are adaptively calculated from the texture density of the image. The border is initially identified by considering the discontinuities in the boundaries. This image is subjected to DBSCAN so that there is no misinterpretation of border points as noise or outlier. The distance function comprises of the seed distance and the neighbour distance components. The pixels in the super-pixel being similar is proved by the seed distance measure and the neighbour distance measure focusses more on the weak boundary and flat regions. The similar neighbouring super pixels are merged to form a single cluster.
Clusters are identified at different densities and according to the data distribution separate Eps and Minpts are considered. If in the adapted Eps and Minpts minimum 10% similar data is available, then a cluster is identified. After 95% of the data are clustered, the remaining points are considered outliers and are discarded. The memory required is O(n2) and the distance matrix size is O(n2 − n)/2 [32]. Instead of reaching a point many times, the computational complexity is minimized by fixing a point to a particular cluster when the adapted Eps and Minpts are optimal. Figure 7 gives the search space of the standard DBSCAN and the proposed approach.
By this approach, linking of clusters by a line, the single link effect is minimized, since arbitrary shaped clusters are permitted. Fractional lp norm distance measure is used instead of Euclidian distance thus minimizing the curse of dimensionality problem [33]. The lp measures identifies if a pixel can belong to a particular cluster forming the local clusters. The similar local clusters are combined to form a global cluster.
5 PSO and GVF Based Segmentation
PSO is an optimization algorithm based on population, modelled according to the behaviour of birds in a flock, identifies global optima by updating the generations [34]. Each particle travels in the space with adjusting the position depending on its own personal best position and the global best by any other swarm, thus reaching global minima faster. The distance from the particle’s present position to the local best position is known as pbest. The distance from the particle’s position to the best global position is gbest. Applying minimum cross entropy thresholding (MCET) as the objective function prevents strucking in sub optima solutions [35, 36]. By this approach, higher dimensional complex particles are reduced to simple one dimensional particles in each space. The pbest of every space is known to every other particles while deciding their pbest using context vector. Figure 8 gives the PSO and adaptive thresholded PSO segmented images and Fig. 9 gives the GVF thresholded segmented image.
The MCET objective function is given by
where j = 1 to M is the histogram of the image, M is the count of gray levels. The optimal threshold value is given by
Thus reducing the computational complexity from \(O(n{L}^{n+1})\) to \(O(n{L}^{2})\). The final context vector is computed by concatenating all the pbest. The final gbest is found analysing all the swarms. Mahalanobis distance measure is used in place of Euclidian distance because each data point is different and may depend on other points also [37]. The squared mahalanobis distance from sample xj and population X is given by
Mahalanobis distance gives dissimilarity between the two random vectors and it is equal to the Euclidean distance if the covariance matrix is identity.
Snake or active contour model with x and y gradient directions with separate force fields for each direction was performed [35]. The angles between the gradients and the contours normal direction at each snake element is used for segmentation. The directional data of the gradient is preserved by the edge map given by
where gx and gy are the smoothed image gradients in horizontal and vertical directions. The external static force from the image and the internal snake force distorts the image by a constant force normal to the image. The elasticity, rigidity, and regularization parameters were fixed to be 0.05, 0 and 0.2 respectively [36, 38]. The segmented images with the combined Affinity propagation and DBSCAN are compared with PSO and GVF segmentation to prove its efficiency in clustering.
6 Dataset and Performance Measures
The proposed hybrid approach was performed on 361 B-mode carotid artery ultrasound images with and without plaque. The images were attained in digital form and discretized to 256 Gy levels. The number of experts for manual segmentation was three. The LI/MA tracings of the image were identified to find the IMT measurement bias to help radiologists do the manual segmentation.
The performance measures used for identifying the optimum segmentation approach for the carotid artery images are as follows.
Intersection over Union J or Jaccard Similarity Coefficient is for comparing the difference and similarity of the sample sets. Its value ranges between 0 and 1. Jaccard distance measures the dissimilarity among the samples and is opposite to jaccard index.
Or it can be written as
where U and V are the two sets, TP is True Positive, TN is True Negative, FP is False Positive and FN is False Negative. The value depends only on the number of pixels correctly and wrongly classified and so it is a gradual measure. Dice index measures the set agreement with the spatial overlap and is a reproducibility variation measure. For the same pair of segmentations dice value is higher and can be used as a lossless function.
The value 1 indicates complete overlap and 0 indicates no overlap of the sets. Hausdorff distance identifies degree to which every point of a model set lies close to certain point of an image set and vice versa. It determines degree of similarity among two objects that are overlaid on one another. It is an indication of the biggest segmentation error. The bidirectional hausdorff distance between the two segmented image sets is given by
where \(hd\left(A,B\right)= {}_{a\in A}{}^{max}{}_{b\in B}{}^{min}{|\left|A-B\right||}_{2}\) and \(hd\left(B,A\right)= {}_{b\in B}{}^{max}{}_{a\in A}{}^{min}{|\left|A-B\right||}_{2}\). Euclidean distance measure is applied here and it gives the maximum distance from a point in one set to the closest point in another set [39].
The accuracy of the segmentation method can be found with probabilistic rand index which gives the similarity between the clusters. It measures the percentage of correct decisions made by the algorithm.
The value close to one indicates a good match with the ground truth segmented image. Negative results are obtained if the segmented results do not match. Cohen’s kappa is a representation of agreement to evaluate the segmentation accuracy, considering that agreement may be just a chance. Kappa value reduces with increasing threshold values. Variation of information (VoI) gives the total information loss and gain among the two clustered results, gives the degree to which one clustering can describe the other. The VOI metric is positive, with smaller values representing better similarity. It indicates the amount of randomness in one segmentation algorithm compared to the other.
where H(A) and H(B) represents the entropy related with the segmented results A and B, and I being the mutual information between A and B. Global consistency error adopts that one segmentation is a modification of the other, and makes all local alterations to be in similar direction. Cophenet measures how loyally a dendrogram conserves the pairwise distances amongst the original unmodeled data points.
7 Results and Discussion
The comparison of the performance of the proposed affinity propagation and DBSCAN with PSO and GVF segmentation approaches is studied in this section. The average of the PSO and GVF segmented results is the ground truth value, which are to be compared with the proposed segmentation approach. Figure 10 gives the edge points extracted by affinity propagation in the sample image.
Wavelet and Ridgelet transforms finds the lines in the image but the curved line segments can be identified only with curvelet transform. The image is divided into smooth overlapping regions and curvelet is applied individually in every region. The method senses anisotropic structures effectively with optimal denoising approximation. The parameters for curvelet thresholding given by Hill et al. are offset parameter α = 1.083, exponent’s scaling product k = 0.790, minimum frequency f0 = 0.509, orientation sub-band frequency offsets g1 = g2 = g3 = 1 [39].
Snake based GVF algorithm has the tendency to converge in places with more luminescence. The dataset is non-linear separable and so the linear discriminant GVF have some errors in concave region. Parameter adjustment and over-segmented region merging because of huge texture variations are done manually. GVF segmented results give good Hausdorff distance and Cohens Kappa values than the PSO and other segmentation methods. During the deformation process, the snakes are re-parameterized to maintain stability. The distance between neighbouring snake contours are maintained at 1 pixel so that the noise remaining does not interfere with the contour. Computation depends on the order of square of the number of contours required making it less time consuming. The snake formation depends on the initial contour selection since it grows from that boundary point. Manual accuracy is an important parameter of the GVF segmentation approach. With its large capture range, the gradient vector gets attracted to the actual boundary and comes closer to that. AN interpolating B spline is created with the obtained continuous contour points.
The global and local optimal values are randomly initialized manually in PSO based segmentation. The computation relies on the population size and is of the order of the product of the count of required iterations and the square of initial population size. On trial 19 iterations gave the best Pbest and Gbest values for most of the carotid artery ultrasound images in the database. The fractional coefficient α is initialized as 1 indicating the non-necessity of memory. This gives pixel wise prediction for deep ensemble base network efficiency. They achieve complementary pixel wise prediction, leading to a smooth contour curve. The residual target echo still attracts to local minima in some images with and without plaque. The inertial weight coefficient φ is made to update in the process to avoid reaching local targets. Initially φ is set to 0.7 and gets updated in each iteration.
Affinity propagation is done with Ramer–Douglas–Peucker algorithm which decimates the segments into similar ones with fewer data points. The first and the last points are kept without alterations. With a threshold considered from the gray histogram, the remaining points are marked as reference to the edge points. Based on the adaptive threshold, the number of data points can be controlled [31, 40, 41]. A mixture density distribution function for l different density types is given by
where wi is the weight of the ith density kind, m is the distance order and \({\mu }_{j}\) is the jth intensity form. Figure 11 gives the DBSCAN clustered image with at distances 32 and 80.
The edge pixels extracted from affinity propagation and the super pixel segmented image are combined and thresholded to obtain the intima-media thickness of the carotid artery. Figure 12 gives the thresholded results of affinity propagation and DBSCAN segmented image.
The mean performance values of the 361 images in the database are compared for the proposed approach combining affinity propagation and DBSCAN with PSO, GVF approaches. The methods are compared with manual segmented results.
Table 1 gives the computational comparison of PSO, GVF and the proposed Affinity propagation and DBSCAN combination algorithms in terms of memory requirement, computational cost, seed points and parameter initializations requirement. Time complexity is a function of the population size and type. Computational complexity involves the generations and evaluations of functions. The cluster parameter α is a damping value between 0.5 and 1. Higher values makes the algorithm converge but increases the computational time. The chunklet data is reduced by a minimizing technique which uses transformation based dimensionality reduction [17].
Similarity metric learning technique is applied to identify and apply only the positive points and to remove the negative points as outliers. The proposed Real time segmentation approach is memory, cost efficient and parameter free in providing the lumen segmentation analysis. The proposed AP + DBSCAN algorithm occupies more memory and is computationally marginally costly. But there is no requirement for initializations of parameters and the cluster count or centroids, making it appropriate for clustering any images. The super pixel segmentation does not require number of iterations to identify the seed points, thus making it a faster clustering approach. The proposed hybrid algorithm runs only once to find the cluster and optimization and so gives good computational performance for real time medical images. Boundary adherence and compact shape is achieved by the affinity propagation algorithm.
The method is fully automatic which do not need ROI initiation and the dataset is optimized for artery distensibility and plaque perfusion assessment. The stiffness parameter of the artery depends on the blood pressure and motion estimation in the artery.
Table 2 compares the segmentation approaches with the manually segmented results, by the clinician’s, in terms of the performance measures. From the experimental results, the proposed approached combining the parameter free affinity propagation and DBSCAN outperforms in terms of Jaccard coefficient, dice index, rand index, variation of information and cophenet. The approach reduces the overall computational complexity, flexibility and gives good error control. The mean intensity and the variance of lumen are less related to its surroundings. The bold values in Table 2 indicates the best performance values. The results shows that the proposed method could consistently track the plaque contour in the carotid artery ultrasound images, which is similar with the manual segmented results.
Localization and segmentation of the carotid artery is a challenging task. The automatic parameter assessment method in the affinity propagation and DBSCAN prevents error while tracking small region of interest. The empirical results state that Affinity propagation with DBSCAN gives impressive performance compared to the traditional PSO, GVF and manual segmentation approaches. The proposed method outperforms the state of art methods in terms of precision, stability and convergence rate. The cooperative method protects the algorithm from falling under curse of dimensionality.
The automatic segmentation approach has a good success rate and also the final results had a very few false positives and false negatives, causing very less number of false diagnosis. The proposed algorithm was designed assuming that the lumen is of smooth curved shape. The mean and variance in intensities of the lumen and the adventitia layers are less compared to the neighbouring layers. The confusion matrix in Table 3 gives the information about the predicted and the true values. The images were obtained with The Ethics Approval Certificate of SRM Medical College Hospital & Research Centre dated 27.06.2019 and numbered 1736/IEC/2019. Toshiba Aplio 400 Ultrasound device was used for ultrasound imaging. The database has 148 cases with disease positive and 198 cases with disease negative.
The proposed approach gives a sensitivity of 93.08%, specificity of 98.01%, precision of 97.36% and accuracy of 95.84%. The confusion matrix is given by
Speckle noise makes the lumen and the hypoechoic tissues similar, which is reduced by curvelet denoising. The outliers in the affinity propagation algorithm are removed by Z score method, where the standard deviations of the points from the mean, assuming Gaussian distribution are calculated and the deviated points are removed. A distinct advantage of our approach is that it is fully user independent and parameter free, making it suitable for segmenting other dataset images. The method provides reliable segmentation of the lumen area in the carotid artery with and without atherosclerosis, which may further used to find plaque perfusion and plaque vulnerability [5].
8 Conclusion
A novel lumen segmentation of the carotid artery combining affinity propagation and DBSCAN clustering approaches is proposed. Affinity propagation identifies outliers, exemplars and clusters which are used to sharpen the DBSCAN clustering. The method is independent of any user defined parameters thus making it suitable for differently oriented carotid artery ultrasound images. Super-pixels are identified and accumulated based on texture and spatial information and are segmented. The algorithm focusses on the overall structure of the image and thus gives global optimal results. The proposed algorithm attains the state–of-the-art performance at much lesser computational and outperforms manual segmentation, GVF snake model and PSO based segmentation with an added parameter free feature. The method is automated, and tried in an extensive set of 361 images and the results are precise. Thus our method can become a valuable technique for carotid artery ultrasound image segmentation. The algorithm focusses on the extracted edge points alone, making it generate larger segments of the lumen region. Future studies with a larger population and combining motion estimation will be performed to improve performance accuracy.
Data Availability
Data available on request from the authors.
Code Availability
Code available on request from the authors.
References
Destrempes, F., Meunier, J., Giroux, M. F., Soulez, G., & Cloutier, G. (2009). Segmentation in ultrasonic B-mode images of healthy carotid arteries using mixtures of nakagami distributions and stochastic optimization. IEEE Transactions on Medical Imaging, 28(2), 215–229.
Hossain, M. M., AlMuhanna, K., Zhao, L., Lal, B. K., & Sikdar, S. (2015). Semiautomatic segmentation of atherosclerotic carotid artery wall volume using 3D ultrasound imaging. Medical Physics, 42(4), 2029–2043.
Ilea, D. E., Duffy, C., Kavanagh, L., Stanton, A., & Whelan, P. F. (2013). Fully automated segmentation and tracking of the intima media thickness in ultrasound video sequences of the common carotid artery. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 60(1), 158–177.
Samiappan, D., & Chakrapani, V. (2016). Classification of carotid artery abnormalities in ultrasound images using an artificial neural classifier. The International Arab Journal of Information Technology, 13(6A), 756–762.
Carvalho, D. D., Akkus, Z., van den Oord, S. C., Schinkel, A. F., van der Steen, A. F., Niessen, W. J., et al. (2015). Lumen segmentation and motion estimation in B-mode and contrast-enhanced ultrasound images of the carotid artery in patients with atherosclerotic plaque. IEEE Transactions on Medical Imaging, 34(4), 983–993.
Jia, D., Cao, J., Song, W. D., Tang, X. L., & Zhu, H. (2016). Colour FAST (CFAST) match: Fast affine template matching for colour images. Electronics Letters, 52(14), 1220–1221.
Wang, X. F., & Huang, D. S. (2009). A novel density-based clustering framework by using level set method. IEEE Transactions on Knowledge and Data Engineering, 21(11), 1515–1531.
Widynski, N., & Mignotte, M. (2014). MultiScale particle filter framework for contour detection. IEEE Transactions on Pattern Analysis and Machine Intelligence, 36(10), 1922.
Yuan, C., Lin, E., Millard, J., & Hwang, J. N. (1999). Closed contour edge detection of blood vessel lumen and outer wall boundaries in black-blood MR images. Magnetic Resonance Imaging, 17(2), 257–266.
Cheng, Y., Hu, X., Wang, J., Wang, Y., & Tamura, S. (2015). Accurate vessel segmentation with constrained B-snake. IEEE Transactions on Image Processing, 24(8), 2440.
Wang, W., Feng, R., Chen, J., Lu, Y., Chen, T., Yu, H., et al. (2019). Nodule-plus R-CNN and deep self-paced active learning for 3D instance segmentation of pulmonary nodules. IEEE Access, 7, 128796–128805.
Arias-Lorza, A. M., Petersen, J., van Engelen, A., Selwaness, M., van der Lugt, A., Niessen, W. J., & de Bruijne, M. (2016). Carotid artery wall segmentation in multispectral MRI by coupled optimal surface graph cuts. IEEE Transactions on Medical Imaging, 35(3), 901–911.
Loizou, C. P., Pattichis, C. S., Pantziaris, M., Kyriacou, E., & Nicolaides, A. (2017). Texture feature variability in ultrasound video of the atherosclerotic carotid plaque. IEEE Journal of Translational Engineering in Health and Medicine, 5, 1–9.
Wang, L., Zhou, X., Xing, Y., Yang, M., & Zhang, C. (2017). Clustering ECG heartbeat using improved semi-supervised affinity propagation. IET Software, 11(5), 207–213.
Li, J. J., Alzami, F., Gong, Y. J., & Yu, Z. (2017). A multi-label learning method using affinity propagation and support vector machine. IEEE Access, 5, 2955–2966.
Zhang, W., Wu, X., Zhu, W. P., & Yu, L. (2017). Unsupervized image clustering with SIFT-based soft-matching affinity propagation. IEEE Signal Processing Letters, 24(4), 461–464.
Yang, C., Bruzzone, L., Zhao, H., Liang, Y., & Guan, R. (2016). Decorrelation–separability-based affinity propagation for semisupervised clustering of hyperspectral images. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 9(2), 568–582.
Yang, C., Bruzzone, L., Guan, R., Lu, L., & Liang, Y. (2013). Incremental and decremental affinity propagation for semisupervised clustering in multispectral images. IEEE Transactions on Geoscience and Remote Sensing, 51(3), 1666.
Yang, C., Bruzzone, L., Sun, F., Lu, L., Guan, R., & Liang, Y. (2010). A fuzzy-statistics-based affinity propagation technique for clustering in multispectral images. IEEE Transactions on Geoscience and Remote Sensing, 48(6), 2647–2659.
Shen, J., Hao, X., Zhiyuan Liang, Yu., Liu, W. W., & Shao, L. (2016). Real-time superpixel segmentation by DBSCAN clustering algorithm. IEEE Transactions on Image Processing, 25(12), 5933–5942.
Hou, J., Gao, H., & Li, X. (2016). DSets-DBSCAN: A parameter-free clustering algorithm. IEEE Transactions on Image Processing, 25(7), 3182.
John, V., Yoneda, K., Liu, Z., & Mita, S. (2015). Saliency map generation by the convolutional neural network for real-time traffic light detection using template matching. IEEE Transactions on Computational Imaging, 1(3), 159–173.
Savaş, S., Topaloğlu, N., Kazcı, Ö., & Koşar, P. N. (2019). Classification of carotid artery intima media thickness ultrasound images with deep learning. Journal of Medical Systems, 43, 273.
Delsanto, S., Molinari, F., Giustetto, P., Liboni, W., Badalamenti, S., & Suri, J. S. (2007). Characterization of a completely user-independent algorithm for carotid artery segmentation in 2-D ultrasound images. IEEE Transactions on Instrumentation and Measurement, 56(4), 1265–1274.
Loizou, C. P., Nicolaides, A., Kyriacou, E., Georghiou, N., Griffin, M., & Pattichis, C. S. (2015). A comparison of ultrasound intima-media thickness measurements of the left and right common carotid artery”. IEEE Journal of Translational Engineering in Health and Medicine, 3, 1–10.
Latha, S., & Samiappan, D. (2019). Despeckling of carotid artery ultrasound images with a calculus approach. Current Medical Imaging, 15(4), 414–426.
Sifakis, E. G., & Golemati, S. (2014). Robust carotid artery recognition in longitudinal B-mode ultrasound images. IEEE Transactions on Image Processing, 23(9), 3762–3772.
Petroudi, S., Loizou, C., Pantziaris, M., & Pattichis, C. (2012). Segmentation of the common carotid intima-media complex in ultrasound images using active contours. IEEE Transactions on Biomedical Engineering, 59(11), 3060–3069.
Panigrahi, S. K., Gupta, S., & Sahu, P. K. (2018). Curvelet-based multiscale denoising using non-local means & guided image filter. IET Image Processing, 12(6), 909–918.
Bao, Q. Z., Gao, J. H., & Chen, W. C. (2008). Local adaptive shrinkage threshold denoising using curvelet coefficients. Electronics Letters, 44(4), 277.
Chakraborty, R., Sushil, R., & Garg, M. L. (2019). An improved PSO-based multilevel image segmentation technique using minimum cross-entropy thresholding. Arabian Journal for Science and Engineering, 44(4), 3005–3020.
Suthar, N., Indr, P., & Vinit, P. (2013). A technical survey on DBSCAN clustering algorithm. International Journal of Scientific & Engineering Research, 4(5), 1775–1781.
Schnitzer, D., & Flexer, A. (2014). Choosing the metric in high-dimensional spaces based on hub analysis. Proceedings of the 22nd European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning, Bruges, Belgium.
Mohsen, F., Hadhoud, M. M., Moustafa, K., & Ameen, K. (2012). A new image segmentation based on particle swarm optimization. The International Arab Journal of Information Technology, 9(5), 487.
Cheng, J., & Foo, S. W. (2006). Dynamic directional gradient vector flow for snakes. IEEE Transactions on Image Processing, 15(6), 1563.
Xu, C., & Prince, J. (1998). Generalized gradient vector flow external forces for active contours. Signal Processing, 71, 131–139.
Zhang, Y., Huang, D., Ji, M., & Xie, F. (2011). Image segmentation using PSO and PCM with Mahalanobis distance”. Expert Systems with Applications, 38, 9036–9040.
Loizou, C. P., Pattichis, C. S., Pantziaris, M., & Nicolaides, A. (2007). An integrated system for the segmentation of atherosclerotic carotid plaque. IEEE Transactions on Information Technology in Biomedicine, 11(6), 661–667.
Hill, P., Achim, A., Al-Mualla, M. E., & Bull, D. (2016). Contrast sensitivity of the wavelet, dual tree complex wavelet, curvelet, and steerable pyramid transforms. IEEE Transactions on Image Processing, 25(6), 2739–2751.
Taha, A. A., & Hanbury, A. (2015). An efficient algorithm for calculating the exact hausdorff distance. IEEE Transactions on Pattern Analysis and Machine Intelligence, 37(11), 2153.
Latha, S., Samiappan, D., & Kumar, R. (2010). Carotid artery ultrasound image analysis: A review of the literature. Part H Journal of Engineering in Medicine, 234, 417.
Acknowledgements
“The authors thankfully acknowledges the financial support provided by The Institution of Engineers (India) for carrying out Research & Development work in this subject”.
Funding
The work is funded by The Institution of Engineers (India).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Ethical Approval
This study and its protocols were approved by SRM Medical College Hospital and Research Centre.
Informed Consent
Informed consent was obtained from all individual participants included in the study. The participant has consented to the submission of the case report to the journal.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Latha, S., Samiappan, D., Muthu, P. et al. Fully Automated Integrated Segmentation of Carotid Artery Ultrasound Images Using DBSCAN and Affinity Propagation. J. Med. Biol. Eng. 41, 260–271 (2021). https://doi.org/10.1007/s40846-020-00586-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-020-00586-9