Abstract
The chemical system for the transformation of energy in eukaryotic mitochondria has engaged researchers for almost a century. This summary of four lectures on the electron transport system in mitochondria is an introduction to the mammalian electron transport chain for those unfamiliar with mitochondrial oxidative phosphorylation. It gives references chosen to reflect the history of the field and to highlight some of the recent advances in bioenergetics. The electron transport chain converts the energy that is released as electrons are passed to carriers of progressively higher redox potential into a proton gradient across the membrane that drives adenosine triphosphate (ATP) synthesis. The electron carriers include flavins, iron–sulfur centers, heme groups, and copper to divide the redox change from reduced nicotinamide adenine dinucleotide (NADH) at −320 mV to oxygen at +800 mV into steps that allow conversion and conservation of the energy released in three major complexes (Complexes I, III, and IV) by moving protons across the mitochondrial inner membrane. The three processes of proton pumping are now known after the successful determination of the structures of the large membrane protein complexes involved. Mitochondria and their proteins play roles not only in the production of ATP but also in cell survival, for which energy supply is the key.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The chemiosmotic mechanism for ATP synthesis is key to aerobic energy conversion in all cells, supplying the majority of the energy required for survival, repair, growth, and reproduction of the organism. There is a wide diversity of electron transport chains across the range of lifeforms, using either light or metabolic energy as the input, with not only oxygen but also other final electron acceptors. These systems not only convert energy from one form (chemical or light) to another (ion gradient across an impermeable membrane and subsequently back to chemical energy in the form of ATP) but also allow the energy to be conserved rather than lost as heat. Bacteria, chloroplasts, and mitochondria transport systems use the energy that is released as electrons are passed to progressively higher redox potential electron carriers to generate proton gradients across membranes that can drive ATP synthesis or transport systems. This article focuses on the components and mechanism of the electron transport chain (ETC) that supports oxidative phosphorylation in mammalian mitochondria, a process described in all biochemistry textbooks, and in more advanced detail in the book Bioenergetics 4 by Nicholls and Ferguson [1].
Figure 1 shows the underlying principle of the chemiosmotic mechanism for energy conversion from its original form to a hydrogen ion gradient that drives the ATP synthase molecular machine. It is the same in chloroplasts using energy from sunlight and in mitochondria using the chemical energy from the breakdown of sugars, proteins, and fats. Although controversial when first described in 1961 and recently republished [2], chemiosmosis was finally accepted as the mechanism enabling oxidative phosphorylation and generation of ATP. Ultimately, Peter Mitchell was awarded a Nobel Prize in 1978 for his innovative work.
The chemiosmotic system requires a proton pump and a membrane to generate a proton gradient and the ATP synthase (ATPase). a Light-driven bacteriorhodopsin incorporated into artificial vesicles with the isolated ATP synthase. Illumination results in a proton gradient that drives ATP synthesis. b The proton pumping complexes (ETC) support the driving force, namely the proton electrochemical gradient Δp, which has two thermodynamically equivalent components: the membrane potential Δψ and the pH gradient (ΔpH)
As shown in Fig. 1, a key experiment proving that a hydrogen ion gradient can drive ATP synthesis was the incorporation of isolated ATP synthase and isolated bacteriorhodopsin into artificial vesicles. Illuminating the bacteriorhodopsin enabled its proton pumping, and ATP was synthesized from adenosine diphosphate (ADP) and phosphate [3, 4]. Just equilibrating isolated mitochondria to pH 8.8 then adding acid to the external medium to give a pH of about 4.3 can generate ATP [5]. Later work showed that movement of organic ions such as succinate could also convert the pH gradient to a membrane potential, and using a bacterial system, that the membrane potential influenced the kinetics of ATP synthase [6]. Figure 1b shows the general scheme with a proton pumping apparatus generating a hydrogen ion gradient across a membrane impermeable to protons. The gradient is then dissipated through the ATP synthase complex, driving conformational changes that are transmitted through the protein to the catalytic trimer of this molecular machine.
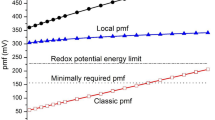
Mitochondria are central to energy integration in eukaryotic cells. Thought to originate from endosymbiosis, each mitochondrion has 5–10 copies of circular DNA and a complete protein translation system in the matrix [7]. The mitochondrial DNA encodes for all the RNA involved in protein synthesis in the matrix but also for 13 proteins that are subunits of the ETC complexes: seven subunits in Complex I, one in Complex III, three in Complex IV, and one in the ATP synthase (sometimes called complex V). To introduce the complexes that will be discussed, Fig. 2 shows a schematic representation of the oxidative phosphorylation system in the inner membrane of mitochondria in mammalian cells. Complexes I and II link metabolic oxidation in the matrix (the citric acid cycle and β-oxidation) to reduction of coenzyme Q (Q, ubiquinone in mammalian systems). Reduced Q (ubiquinol, QH2) is reoxidized by Complex III that passes electrons via cytochrome c to Complex IV, the terminal oxidase. Electron transfer from a low redox potential donor to a higher redox potential acceptor in three complexes is coupled to proton movement from the matrix to the intermembrane space (IMS) that equilibrates with the cytosol of the cell. These three complexes (I, III, and IV) are sometimes called the “coupling sites” of the respiratory chain. The proton gradient (with the thermodynamically equivalent membrane potential) across the membrane drives ATP synthesis via the ATP synthase in intact coupled mitochondria. In the steady state, respiring mitochondria support a transmembrane electrochemical gradient of around 220 mV. The proton motive force can be expressed as
where Δψ is the membrane potential and ΔpH is the pH difference across the membrane, both expressed as the cytoplasmic minus the matrix values. The membrane potential is positive outside and the pH is lower outside than in the matrix.
Mitochondrial oxidative phosphorylation. a Components of the electron transport system from NADH to oxygen that transfer protons from inside to outside the inner membrane: Complexes I, II, and IV. The F1F0ATP synthase (complex V) allows proton translocation from outside the inner membrane (high H+ concentration) to the matrix (low H+) to drive ATP synthesis. b Oxygen consumption by mammalian mitochondria in the presence of pyruvate and malate that support generation of NADH in the matrix. The slow rate observed at the start of each trace is called state 2 (substrate but no ADP). When uncoupler is added (left), the maximum rate of the ETC is observed as the system is no longer limited by a high electrochemical potential. When ADP is added (right), the ATP synthase facilitates proton movement into the matrix (state 3 rate of oxygen consumption) until all the ADP has been converted to ATP, when the rate is again limited by the high electrochemical potential (state 4 rate)
Figure 2 also shows the consumption of dioxygen by mitochondria, a measure of the rate of electron transport. If mitochondria are uncoupled by breaking the membrane or by adding an uncoupler (a membrane-permeant weak acid that acts to transfer protons from outside to the matrix), then the ETC can work at its maximum rate (Fig. 2b, left). Healthy mitochondria have a proton-impermeable membrane that supports the proton gradient. These mitochondria are termed “coupled” because the passage of electrons along the ETC (and hence the consumption of dioxygen) is coupled to ATP synthesis. In coupled mitochondria, the maximum rate of ETC, measured as oxygen consumption, is only observed in the presence of ADP (Fig. 2b, right). The respiratory control ratio (RCR) is used to define coupling: it is the ratio of the rate of oxygen consumption in the presence of high ADP to the rate in the presence of high ATP. Lower values of RCR indicate that protons leak through the membrane, when the energy is released as heat rather than conserved as ATP.
This article describes the electron transport system and how the movement of electrons is coupled to proton pumping to generate the hydrogen gradient across the mitochondrial inner membrane. The release of energy from electrons passing along electron carriers from lower to higher potential drives proton pumping in the three key membrane complexes of the electron transport chain, viz. Complex I, Complex III, and Complex IV (Fig. 2). Enormous crystallography challenges have been overcome to provide the structures of the complexes. When combined with functional studies using spectroscopy and electron paramagnetic resonance (EPR) and a wealth of information from bacterial and plant systems, these structures support a detailed picture of the fascinating mitochondrial energy-transforming machinery.
Cofactor structures and redox potentials
The redox cofactors are held within the proteins that provide insulation from the surrounding aqueous environment as well as binding sites for the chemicals required to donate and accept electrons. The mammalian electron transport chain (ETC) contains the flavins, iron–sulfur clusters (Fe–S), several types of heme, and, in the terminal oxidase, copper (Cu), structures for which are shown in Figs. 3 and 4. The families of redox carriers have different properties that come from variants of cofactor structure and from the specific protein environment, as will become apparent from the brief description here and can be found in more depth in the references.

(Adapted from [12] with permission from American Chemical Society Publications)
Redox carriers in cytochrome oxidase (Complex IV). The binuclear CuA center (brown balls; the ligands are detailed in the box on the right) accepts an electron from cytochrome c (red circle). Electrons pass to the two heme groups buried in the helices, cytochrome a (LS, low-spin heme in the bottom box) and cytochrome a3 (HS, high-spin heme), and to the CuB that binds to dioxygen.
Flavins Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), derived from the vitamin riboflavin in humans, can be reduced by one electron to the flavosemiquinone, or by two electrons to the flavohydroquinone (Fig. 3). All forms can be protonated or not depending on the environment [8]. The redox potentials for the two-electron reduction lie in the region of −0.2 V, but the exact redox potential is modulated by the protein: for example, the covalently bound FAD in beef heart Complex II (succinate dehydrogenase) has a midpoint potential of −0.079 V. This also links to catalysis: electrocatalytic voltammetry studies demonstrated that redox poise and pH strongly influence the catalytic activity of succinate dehydrogenase, favoring succinate oxidation only above pH 7.7 [9].
In the ETC, Complex I (NADH-Q oxidoreductase) contains FMN, whereas FAD is the cofactor in the non-proton pumping enzymes, succinate dehydrogenase (succinate-Q oxidoreductase, Complex II) in the citric acid cycle, and electron-transferring flavoprotein (ETF)-Q oxidoreductase that brings electrons from fatty acid oxidation. The role of the flavins in Complexes I and II is to transduce the two-electron input from NADH or succinate, respectively, to pass one electron down the subsequent series of one-electron-carrying iron–sulfur clusters in these complexes.
Iron–sulfur clusters The mitochondrial respiratory chain contains clusters with two, three, or four iron atoms bridged by sulfur atoms. The 2Fe–2S and 4Fe–4S clusters shown in Fig. 3 are liganded to the protein via cysteine residues. Each cluster is reduced by only one electron, but the charge is unevenly distributed over the metal atoms (a mixed valence state). Reduced forms are detectable by electron paramagnetic resonance (EPR) (S = ½ for most of the ETC clusters). The EPR signals provided a useful tool to determine reduction in the mitochondrial membrane in the 1960s (reviewed in [10]), and EPR continues to be used to examine the various proportions of oxidized and reduced carriers (their redox status) in complex systems [11].
Heme groups Cytochromes contain heme groups: iron surrounded by a porphyrin ring that provides four of the coordination sites, with the fifth and sixth coordination provided by amino acid side-chains, usually histidine or methionine. The different types of heme are characterized by distinctive visible spectral changes on reduction; for example, the difference maxima for the two b-type cytochromes (b556 and b564) in Complex III are 556 nm and 564 nm. These two b cytochromes are also distinguished by electron paramagnetic resonance (EPR) as bL and bH, indicating low- and high-spin behavior, respectively. Hemes are single-electron carriers. Those found in the mammalian mitochondrial ETC have redox potentials above 0 mV (Table 1), with the potential depending upon the substitutions on or puckering of the porphyrin ring; For example, cytochromes with methionine coordinating to the iron at the axial positions have potentials about 100 mV higher than those with histidine. In general terms, the redox potential is influenced by solvent exposure, by electrostatic interactions inside the protein, and by the axial ligands.
The cytochrome heme groups function to carry electrons from the mobile coenzyme ubiquinol (reduced from Q to QH2 by Complexes I and II as described below) to dioxygen. Figure 3c shows a c-type heme such as that found in cytochrome c, the mobile carrier between Complex III and Complex IV. The c-type heme in cytochrome c, a small highly conserved protein of only about 12 kDa, has histidine as the fifth coordination site ligand to the iron atom and methionine as the sixth. To facilitate electron transfer, the cytochrome c binds to Complex III to accept an electron and to Complex IV to donate an electron via electrostatic binding, which results in the donating and accepting hemes being positioned close together and away from bulk solvent.
Copper centers Heme groups carry only one electron, but four electrons are required to reduce dioxygen to water at the end of the respiratory chain. This is achieved in Complex IV (cytochrome oxidase) using a combination of hemes and copper centers [binuclear copper-A (CuA), cytochrome a, cytochrome a3, and copper-B] (Fig. 4). The CuA center has two coppers with histidine and methionine and a main-chain carbonyl as ligands. Although there are two coppers, the CuA center accepts only one electron, which is delocalized across both coppers.
Major complexes and mobile carriers
The electron carriers are reduced according to the generalized half-reaction
where n is the number of electrons involved and m is the number of hydrogens. The midpoint potential is the potential at which half is oxidized and half is reduced (see [1, Chap. 3] for a full explanation). The midpoint redox potentials of the electron carriers (Table 1) feed into the thermodynamic prediction of their order in the electron transport chain order, but it is not that simple. The relationship between the Gibbs free energy for electron transfer between two redox couples at pH 7 is calculated from the redox potential difference for their oxidation/reduction. For each of two components A and B:
where n is the number of electrons transferred, R is the gas constant, F is the Faraday constant, Eh is the midpoint redox potential of the half-reaction, and ΔEh is the difference between the two redox potentials. In biological systems, the values are given for pH 7. Where protons are involved in a reaction, the pH changes the concentration-dependent redox potential of the half-reaction,
Considering the whole electron transport chain, the electron transfer from NADH (Eh = −320 mV) to quinone (Eh = +4 mV) under standard conditions at pH 7 would be associated with ΔG = −2 (0.0965 kJ mol−1 mV−1) (4 − (−320) mV) = −62.5 kJ mol−1, being strongly energetically favorable and driven by a constant supply of NADH from cellular metabolism via the citric acid cycle (i.e., the system is far from equilibrium). It is important to note that, under physiological conditions, the proportions of oxidized and reduced components vary according to the metabolic conditions and the availability of oxygen, so ΔG values must be calculated from concentration-dependent potentials. Thus, reverse electron transport from Q to NADH is possible when the concentration of succinate is high and that of dioxygen is low. In addition to alterations in the concentration of reductants, the redox potential difference is modulated if the donor and acceptor couples are on the opposite sides of a membrane where there is a transmembrane potential, as exists across the mitochondrial inner membrane. The three complexes associated with electron transfer that drive proton movement across the membrane all have a substantial potential difference between the donor and acceptor (NADH and Q, respectively, in the example above). In contrast, the ΔEh for succinate to Q is close to zero, so that no work can be done—there is no proton transfer by Complex II.
So, how was the order of the complexes shown in Fig. 5a and their electron carriers discovered? Early work on beef heart mitochondria, summarized by Ernster in 1981 [14], led to the isolation of the complexes and characterization of the cofactors in the 1950s. Key tools were inhibitors selective for each complex: rotenone or piericidin for Complex I, thenoyltrifluoroacetone (TTFA) or carboxins for Complex II, antimycin or myxothiazol for Complex III, and cyanide or carbon monoxide for Complex IV (Fig. 5a). These chemicals are still used in mitochondrial experiments today, as shown in the typical dioxygen electrode trace in Fig. 5b. Establishing the order of the carriers required painstaking analysis of visible spectra (see Fig. 5c) and electron paramagnetic signals at poised potentials and their changes with time after inhibition or varying dioxygen. When dioxygen was introduced to the fully reduced ETC, the order in which the cofactors reoxidized helped verify their order. Time-resolved spectroscopy has been used in modern format to investigate supercomplexes within the ETC [15].
Demonstrating the order of electron transport complexes. a ETC showing substrates and inhibitors. Q in mammalian mitochondria is ubiquinone. b O2 electrode trace for uncoupled mitochondria, showing effects of adding substrates and inhibitors in sequence. N,N,N′,N′-Tetramethyl-p-phenylenediamine (TMPD) mediates electron flow from ascorbate to Complex IV. c Visible spectral changes after CO inhibition at cytochrome a3 reproduced from [15] with permission [copyright (2008) National Academy of Sciences]
The organization of the electron carriers within each of the complexes was only fully appreciated after the determination of crystal structures from the 1990s onwards. Complete structures were determined for Complex II with only four subunits (reviewed in [16]), followed by Complex III with 11 subunits [17] and Complex IV with 13 subunits [18]. Complex I, with more than 45 subunits in its mammalian form, many of them highly hydrophobic, was the greatest challenge (see below), with only shapes based on electron microscopy available at the turn of this century. The electron pathways within each complex are shown in Fig. 6.
Organization of cofactors in the complexes and the structures of the mobile carriers. CI. Arrangement of FMN and 4Fe–4S and 2Fe–2S clusters in the peripheral arm of Complex I from Yarrowia lipolytica [20], showing center-to-center (and edge-to-edge in brackets) distances between the electron carriers in angstroms. CII. Distances between FAD, Fe–S centers, and Q (labeled UQ) in Complex II. CIII. Distances between the electron carriers (Fe–S and heme) from Q to cytochrome c in the Complex III dimer. (Reproduced from [23] with permission, copyright (2007) National Academy of Sciences USA.) The round circles highlight the pair-splitting site, the ovals the semiquinone stabilization sites. CIV. Cartoon of the arrangement of electron carriers in Complex IV (reproduced from [24], with permission, copyright (2008) National Academy of Sciences USA)
Complex I NADH-Q oxidoreductase is an essential component of energy transformations in all species, with 14 core subunits conserved from bacteria to primates. The crystal structure at 6.3 Å of the eukaryotic mitochondrial Complex I from Yarrowia lipolytica [19, 20] with 40 subunits (giving a total molecular weight of 946.5 kDa) revealed a boot-shaped structure running about 180 Å along the membrane with the “leg” protruding into the mitochondrial matrix. It is in this protruding part that the electron carriers of the almost 100-Å-long electron transport pathway are found, with the binding site for Q buried in the protein on the matrix side of the membrane. The crystal structure of 16 subunits of Complex I from Thermus thermophilus, at 3.3 Å resolution [21], revealed 64 transmembrane helices and 9 iron–sulfur clusters (Fig. 6, top left). This structure enabled a detailed proposal for redox-driven proton pumping that is described below. The first mammalian structure at 5 Å derived from cryoEM and modeling [22] was for the beef mitochondrial Complex I with 45 subunits and 8 iron–sulfur centers, allowing correlation with earlier functional and EPR studies that used Complex I purified from beef tissues.
Complex II Succinate-Q oxidoreductase has two peripheral subunits containing the electron carriers and two transmembrane subunits essential for reduction of Q (denoted as UQ in Fig. 6). The electron transport pathway (Fig. 6, top right) spans 50 Å, but as in Complex I, the distances between each carrier are short enough to allow rapid electron transfer. A review of the structure and function of succinate dehydrogenases and the closely related bacterial fumarate reductases gives a full description of the experiments leading to an understanding of the function [16].
Coenzyme Q Complexes I and II and other redox enzymes, such as ETF-Q oxidoreductase which accepts electrons from fatty acid metabolism, reduce Q (ubiquinone/ubiquinol, midpoint potential +40 mV, n = 2). Many bacteria use menaquinone, and photosynthesis in plants uses plastoquinone, variants on the mammalian ubiquinone (Q) shown in Fig. 6. Note the long (50 carbon) hydrophobic tail which keeps the Q in the membrane. If concentrations of Q in the membrane decrease, net electron transport is decreased.
Complex III Complex III from bovine heart was crystallized as a dimer of 2 × 240 kDa with 11 subunits in each monomer [17]. The first electron acceptor from the reduced mobile carrier ubiquinol (QH2) is a 2Fe–2S center (+280 mV) in the Rieske complex that protrudes into the IMS. The 2Fe–2S center passes the electron to cytochrome c1, from where the electron exits Complex III to the mobile cytochrome c. However, QH2 is a two-electron donor, so Complex III oxidizes QH2 at a pair-splitting Q-site that gives concerted one-electron reduction of the 2Fe–2S center (+280 mV) and of heme BL (−90 mV) with loss of two H+ to the IMS. The latter electron passes rapidly (400 s−1) from heme BL to heme BH (+40 mV), where it is used to reduce Q to the semiquinone in the stabilization site. Stabilization of the resulting anionic semiquinone is achieved by a hydrogen bond to histidine and hydrophobic groups stacked above and below the quinone ring. The process is repeated with a second molecule of QH2, resulting in two-electron reduced Q in the stabilization site. The process is known as the Q-cycle and results in transfer of protons across the membrane (see below).
In the Q-cycle, two H+ are taken up from the matrix to give QH2, which is released. Meanwhile, each electron passed to the Rieske 2Fe–2S moves to the cytochrome c1 (+240 mV at 164 s−1) and then to the mobile carrier, cytochrome c. Thus, although two QH2 are oxidized, one QH2 is produced in the cycle, so there is net oxidation of one QH2 and reduction of two molecules of cytochrome c. Accompanying the electron transfer, two protons are absorbed from the matrix and four protons are released into the matrix (see below). The arrangement of the carriers and the two Q-sites is shown in Fig. 6 (bottom left) [23]. Both hemes are in the transmembrane helical subunits, while the Rieske 2Fe–2S and cytochrome c1 are located on the outer part of the membrane. The iron to cytochrome c1 distance of 31.6 Å is decreased to 21.3 Å when the 2Fe–2S center is reduced, thus decreasing the distance for onward flow of electrons.
Cytochrome c The mobile carrier linking Complexes III and IV is the small (12 kDa) highly conserved protein cytochrome c (Fig. 6, ribbon diagram). The heme group (+254 mV) is attached to the protein motif CXXCH via two thioether bonds, with one edge of the porphyrin ring exposed to the solvent. This edge is presented to the binding partners for electron transfer thanks to electrostatic attraction between surface lysine residues on cytochrome c and spaced acidic residues on the donor and acceptor complexes. Cytochrome c is located on the outer face of the inner membrane in the intermembrane space. If the outer membrane of the mitochondria is ruptured, cytochrome c escapes into the cytosol. Not only does this decrease the rate of electron transport but it also activates caspases, degradative enzymes in the cytosol, and acts as a trigger for apoptosis.
Complex IV The seven-subunit cytochrome oxidase accepts four electrons in turn from cytochrome c to reduce dioxygen to water. The electron pathway (Fig. 6, bottom right) is from cytochrome c to the binuclear copper A center to cytochrome a, to the cytochrome a3 adjacent to copper-B, about 1/3 of the way into the membrane from the IMS. When cytochrome a3 is reduced, dioxygen can bind to be reduced to peroxide and then to water (see [25] for the proposed mechanism) with concomitant pumping of protons.
Electron transfer rates Electron transfer must be efficient to maximize energy conservation and to support the rapid rate of ATP supply required by the organism. Direct electron transfer from the low-potential carriers to dioxygen would not only generate heat rather than conserving energy but would also generate reactive dioxygen species deleterious to the cell. This “leakage” of electrons is minimized by the location of electron carriers inside the proteins, as seen in the deep Q-sites or in the tightly oriented pairing of cytochrome c with its donor and acceptor complexes. The distance between electron carriers is important for the rate of electron transfer, with the rate decreasing roughly 106-fold per 10 Å (see [26] for the theory supported by experimental observations). Looking at the distances in Fig. 5, the rates of electron transfer in the ETC will be around 106 s−1 for the 15 Å prevalent in Complex I. In Complex III, although the distance of 32 Å from the oxidized Rieske center to cytochrome c1 would predict a rate of less than 1 s−1, the movement of the Fe–S center closer to cytochrome c1 upon reduction results in a distance of 21 Å, enabling electron transfer at a rate consistent with the rest of the ETC.
Recent studies on the complexes indicate close association of the complexes in the mitochondrial cristae (reviewed in [27]). Complex I forms a supercomplex with Complexes III and IV, known as the respirasome, thought to allow more efficient electron transfer by minimizing the distance traveled by the mobile carriers.
Proton pumping
Each of the three complexes of the pathway from NADH to dioxygen has a different mechanism for converting the energy from electron transfer into a hydrogen ion gradient across the mitochondrial inner membrane (Fig. 7). Complex I has four antiporter-like channels opened by conformational change linked to electron transfer by the redox state of quinone [19, 21, 28]. In Complex III, the Q-cycle releases four protons from the oxidation of 2QH2 in the pair-splitting site into the IMS, and the concomitant protonation of Q being reduced at the stabilization site uses two protons from the matrix [29]. Complex IV has proton relay channels lined with acidic amino acids [30,31,32].
Three ways to pump protons. a Complex I: four-channel type opened by conformational change linked to electron transfer by the redox state of quinone. Adapted from Baradaran et al. [21] with permission from Nature, copyright (2013). b Complex III: In the Q-cycle, 4e− from 2QH2 are split to reduce two cytochrome c in the IMS, and the other 2e− are delivered via cytochromes bL and bH to reduce Q at the matrix face of the membrane. The four protons from the 2QH2 oxidized at the IMS surface of the membrane are released into the IMS, while the Q reduced at the matrix face obtains two protons from the matrix. c Complex IV: Proton channels (H +D and H +K ) conduct protons for the reduction of dioxygen and to a “proton-loading site” for discharge to the outside [30]
The stoichiometry of protons translocated to electrons (nH+/2e− stoichiometry) passing down the chain has been much studied. The experiments using dyes, pH electrodes, and the ratio of the amount of dioxygen consumed to the amount of ADP phosphorylated are difficult, confounded by proton leak and experimental limitations that contribute to uncertainty [33]. For the individual complexes, nH+/2e− stoichiometry can be measured using artificial electron acceptors to measure the electrons and using potassium ions in the presence of the ionophore valinomycin to quantify the charge movement, interpreting results in the light of structural knowledge. The value for Complex I is 4H+/2e−, recently confirmed using a combination of experiment and theory [34]. Complex III releases 4H+ from 2QH2 oxidized at the pair-splitting site, with 2e− passing to cytochrome c. The other two electrons are passed to the b-type hemes to reduce Q at the semiquinone stabilizing site, where 2H+ are taken from the matrix. Complex IV requires two scalar protons for each water formed but also pumps 2H+/2e−. For the full ETC, a stoichiometry of 10H+/2e− for NADH to dioxygen and of 6H+/2e− for succinate to dioxygen are generally accepted (see Nicholls & Ferguson, Bioenergetics 4 [1]). On the side using the electrochemical gradient to make ATP, the mammalian stoichiometry is 2.67H+/ATP [35].
Finally, note that not all the energy is converted to ATP, but instead the electrochemical gradient is applied directly to work for the cell homeostasis. The electrochemical gradient drives the export of ATP from the mitochondria, the import of ADP, phosphate, and other small molecules, and the accumulation of calcium ions, and is required for the import of proteins. If the membrane impermeability to hydrogen ions is breached by the addition of an uncoupler such as dinitrophenol (DNP), the hydrogen ion gradient collapses and the ETC works at maximum rate without generating ATP but producing heat instead. The mitochondrial inner membrane does contain special proteins that can allow small rates of leak. These uncoupling proteins are found mainly in brown fat, where they allow the production of heat especially in neonates and young animals [36].
Inhibition
Inhibitors acting specifically at the various Q-sites in the ETC are indicated in Fig. 5, and some of their structures are shown in Fig. 8. In 1998, Davey et al. explored how much each complex had to be inhibited in order to see an effect on ATP generation or dioxygen consumption [37]. Complex I has the least spare capacity, because decreased ATP formation and dioxygen consumption was seen with only 30% inhibition by rotenone. In contrast, Complex III had to be inhibited by more than 80% with antimycin A, and complex IV by more than 75% with cyanide, before the rate of dioxygen consumption decreased. The low threshold for Complex I means that even slight inhibition can be deleterious to the energy supply of the cell.
The smallest structure in Fig. 8 is 1-methyl-4-phenylpyridinium (MPP+), which binds at the Q-sites of complex I. Produced in the brain as the oxidation product from the neurotoxin, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), MPP+ is accumulated into dopaminergic neurons by the dopamine reuptake carrier and then again accumulated as a result of its positive charge (a delocalized charge that does not prevent membrane penetration) into the mitochondrial matrix. The amount of accumulation depends on the membrane potential, so that a 50 μM cytosolic concentration could give as much as 20 mM in the matrix [38]. MPP+ inhibits at two sites on complex 1, preventing electron transport, stopping production of ATP (without which the cell dies), and resulting in drug-induced Parkinsonism [39]. Rotenone acts at the same site and is used as a pesticide to kill insects on plant crops.
Leak of electrons from the ETC to dioxygen
The electron carriers in the ETC are well insulated by the proteins around them, even at the Q sites, which are deep within the structures. Nonetheless, electrons can be passed from the ETC to dioxygen to generate the superoxide radical (O ·−2 , which collects a proton at cellular pH to give HO ·2 ). Superoxide, able to cross membranes only very slowly, is released into the matrix or cytosol, where it has signaling roles. The cell has strong defenses to protect against deleterious elevated levels. The superoxide dismutases are copper/zinc (cytosol) or iron/manganese (mitochondria) containing enzymes that dismutate superoxide into the less reactive (but membrane permeant) hydrogen peroxide (H2O2) that is converted into water and dioxygen by catalase. The tripeptide antioxidant glutathione is another major defense, protecting key thiols in proteins, maintained in reduced state by glutathione reductase. The levels of glutathione in cells are used as a measure of oxidative stress—glutathione depletion means that the cell has excess reactive dioxygen species (ROS, shown in Fig. 9a) that damage lipids, proteins, and nucleic acids, damage which can eventually cause cell death.

(reprinted from TIBS [43] with permission from Elsevier)
ROS species, their generation, and the sources of superoxide formation in the ETC. a ROS and the redox potentials for interconversion. b Haber–Weiss and Fenton reactions generate hydroxyl radical. c Sites of superoxide formation in the ETC
As well as superoxide and hydrogen peroxide (Fig. 9a), the highly reactive hydroxyl radical is a potent source of damage to biomolecules, attacking, for example, the double bond in fatty acids in the membranes to form peroxides. The generation of hydroxyl radicals occurs in the cell in the presence of reduced metal ions, particularly the ubiquitous ferrous iron (FeII). The Haber–Weiss reaction uses hydrogen peroxide and superoxide; the Fenton reaction generates the hydroxyl radical from hydrogen peroxide alone (Fig. 9b).
The ETC is the major source of superoxide in mammalian cells, mainly from the Q-sites (Fig. 9c). Some ROS are also generated from the flavin sites of several dehydrogenases (for example, Complex I, Complex II, ETF-Q oxidoreductase, and pyruvate dehydrogenase) [40]. The leak of electrons from Q-sites to dioxygen in Complex I is likely to come from the semiquinone moiety formed by one-electron transfer from the last carrier in Complex I (Fe–S center N2). In Complex III, the major site is the outer, pair-splitting site Qo, located near cytochrome b556 and blocked by myxothiazol. This site releases the superoxide to the cytosol. The rate of generation of ROS depends on the redox status of the ETC (recently reviewed [40]). The capacity for ROS generation is greatest from Complex III, then Complex I, with the Complex II flavin contributing much less [41]. However, the quantities vary with redox state [42]. Under normal working conditions in muscle mitochondria, less than 1% of the dioxygen turnover in the ETC results in ROS.
Cell-based bioenergetics and concluding remarks
This article has presented an overview of the chemistry of the ETC in mammalian systems. To finish, I would like to point readers also to the wide diversity of ETCs across living systems introduced briefly with further reading in [1], and to the surge in medical research publications reporting measurement of bioenergetics parameters in mammalian cells.
The ability to measure bioenergetics parameters in cells depends on sensitive measurement of dioxygen consumption before and after carefully defined addition of first an ATPase inhibitor (oligomycin) to define ATP production in the respiring cells, then an uncoupler [carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP)] to allow maximal respiration, then inhibitors (rotenone and antimycin A), as shown in Fig. 10. A good description of the method can be found in [44], and detailed information and webinars about the techniques used in the Agilent Seahorse XF Analyzers are available at the Agilent website. The methodology has become a key technique for assessment of cell life and death, for cancer treatment development, and for understanding the complexities of metabolic regulation.

(Figure from Agilent Technologies Seahorse XF assay, https://www.agilent.com/)
Determining bioenergetics parameters for cells. Live cultured cells are incubated in a closed chamber to measure the oxygen consumption. Addition of the ATPase inhibitor oligomycin stops ATP production in the respiring cells, leaving the oxygen consumption rate due to proton leak plus the nonmitochondrial consumption. Addition of the uncoupler FCCP allows maximal respiration, then inhibitors (rotenone and antimycin A) to completely block mitochondrial oxygen use and take the oxygen consumption back down to nonmitochondrial use only.
Bioenergetics are key to cell survival. If ATP production is compromised, both signaling and damage alter homeostasis in the cell. Mitochondria and their proteins play a key role not only in the production of ATP but also in cell survival. The description of the ETC presented here provides a simple basis for further exploration of the mechanism of energy transduction and cell responses to changes in energy.
References
Nicholls DG, Ferguson SJ (2013) Bioenergetics 4. Academic, London
Mitchell P (2011) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biochim Biophys Acta Bioenerg 1807:1507–1538
Winget GD, Kanner N, Racker E (1977) Formation of ATP by adenosine-triphosphatase complex from spinach-chloroplasts reconstituted together with bacteriorhodopsin. Biochem Biophys Acta 460:490–499
Oren R, Weiss S, Garty H et al (1980) ATP synthesis catalyzed by the ATPase complex from rhodospirillum-rubrum reconstituted into phospholipid-vesicles together with bacteriorhodopsin. Arch Biochem Biophys 205:503–509
Reid RA, Moyle J, Mitchell P (1966) Synthesis of adenosine triphosphate by a protonmotive force in rat liver mitochondria. Nature 212:257–258
Kaim G, Dimroth P (1999) ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J 18:4118–4127
Gustafsson CM, Falkenberg M, Larsson NG (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem 85:133–160
Entsch B, Ballou DP (2013) Flavins. In: Lennarz WJ, Lane MD (eds) Encyclopedia of Biological Chemistry. Academic Press, London, pp 309–313
Hirst J, Sucheta A, Ackrell BAC et al (1996) Electrocatalytic voltammetry of succinate dehydrogenase: direct quantification of the catalytic properties of a complex electron-transport enzyme. J Am Chem Soc 118:5031–5038
Beinert H (1992) Trails of inquiry and thought leading toward todays bioenergetics. Biochem Biophys Acta 1101:125–133
Hagen WR (2018) EPR spectroscopy of complex biological iron–sulfur systems. J Biol Inorg Chem 23:623–634
Liu J, Chakraborty S, Hosseinzadeh P et al (2014) Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem Rev 114:4366–4469
Vos MH, Liebl U (2015) Time-resolved infrared spectroscopic studies of ligand dynamics in the active site from cytochrome c oxidase. Biochim Biophys Acta Bioenerg 1847:79–85
Ernster L, Schatz G (1981) Mitochondria—a historical review. J Cell Biol 91:S227–S255
Trouillard M, Meunier B, Rappaport F (2011) Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc Natl Acad Sci USA 108:E1027–E1034
Cecchini G (2003) Function and structure of Complex II of the respiratory chain. Annu Rev Biochem 72:77–109
Iwata S, Lee JW, Okada K et al (1998) Complete structure of the 11-subunit bovine mitochondrial cytochrome bc(1) complex. Science 281:64–71
Tsukihara T, Aoyama H, Yamashita E et al (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 angstrom. Science 272:1136–1144
Hunte C, Zickermann V, Brandt U (2010) Functional modules and structural basis of conformational coupling in mitochondrial Complex I. Science 329:448–451
Zickermann V, Wirth C, Nasiri H et al (2015) Mechanistic insight from the crystal structure of mitochondrial Complex I. Science 347:44–49
Baradaran R, Berrisford JM, Minhas GS et al (2013) Crystal structure of the entire respiratory Complex I. Nature 494:443–448
Vinothkumar KR, Zhu JP, Hirst J (2014) Architecture of mammalian respiratory Complex I. Nature 515:80–84
Zhu J, Egawa T, Yeh SR et al (2007) Simultaneous reduction of iron–sulfur protein and cytochrome b(L) during ubiquinol oxidation in cytochrome bc(1) complex. Proc Natl Acad Sci USA 104:4864–4869
Gorbikova EA, Belevich I, Wikstrom M et al (2008) The proton donor for O–O bond scission by cytochrome c oxidase. Proc Natl Acad Sci USA 105:10733–10737
Wikstrom M (2012) Active site intermediates in the reduction of O-2 by cytochrome oxidase, and their derivatives. Biochim Biophys Acta Bioenerg 1817:468–475
Moser CC, Keske JM, Warncke K et al (1992) Nature of biological electron-transfer. Nature 355:796–802
Enriquez JA (2016) Supramolecular organization of respiratory complexes. Annu Rev Physiol 78:533–561
Kaila VRI (2018) Long-range proton-coupled electron transfer in biological energy conversion: towards mechanistic understanding of respiratory complex I. J R Soc Interface 15:20170916
Crofts AR (2004) The cytochrome bc(1) complex: function in the context of structure. Annu Rev Physiol 66:689–733
Belevich I, Bloch DA, Belevich N et al (2007) Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc Natl Acad Sci USA 104:2685–2690
Rich PR, Marechal A (2013) Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J R Soc Interface 10:20130183
Wikstrom M, Sharma V, Kaila VR et al (2015) New perspectives on proton pumping in cellular respiration. Chem Rev 115:2196–2221
Ferguson SJ (2010) ATP synthase: from sequence to ring size to the P/O ratio. Proc Natl Acad Sci USA 107:16755–16756
Jones AJY, Blaza JN, Varghese F et al (2017) Respiratory Complex I in Bos taurus and Paracoccus denitrificans pumps four protons across the membrane for every NADH oxidized. J Biol Chem 292:4987–4995
Silverstein TP (2014) An exploration of how the thermodynamic efficiency of bioenergetic membrane systems varies with c-subunit stoichiometry of F1F0 ATP synthases. J Bioenerg Biomembr 46:229–241
Bertholet AM, Kirichok Y (2017) UCP1: a transporter for H+ and fatty acid anions. Biochimie 134:28–34
Davey GP, Peuchen S, Clark JB (1998) Energy thresholds in brain mitochondria—potential involvement in neurodegeneration. J Biol Chem 273:12753–12757
Ramsay RR, Singer TP (1986) Energy-dependent uptake of N-methyl-4-phenylpyridinium, the neurotoxic metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, by mitochondria. J Biol Chem 261:7585–7587
Przedborski S, Tieu K, Perier C et al (2004) MPTP as a mitochondrial neurotoxic model of Parkinson’s disease. J Bioenerg Biomembr 36:375–379
Brand MD (2016) Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radical Biol Med 100:14–31
Quinlan CL, Perevoshchikova IV, Hey-Mogensen M et al (2013) Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol 1:304–312
Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417:1–13
Hamanaka RB, Chandel NS (2010) Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci 35:505–513
Brand MD, Nicholls DG (2011) Assessing mitochondrial dysfunction in cells. Biochem J 435:297–312
Acknowledgements
This article is dedicated to the many students who have shared my enthusiasm for mitochondria in lectures and practical classes for the last 24 years and to my colleagues in mitochondrial research. I thank especially the students this year who gave me feedback on the article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ramsay, R.R. Electron carriers and energy conservation in mitochondrial respiration. ChemTexts 5, 9 (2019). https://doi.org/10.1007/s40828-019-0085-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40828-019-0085-4