Abstract
Polarized and wide-field light microscopy has been studied for many years to develop accurate and information-rich images within a focused framework on biophysics and biomedicine. Technological advances and conceptual understanding have recently led to significant results in terms of applications. Simultaneously, developments in label-free methods are opening a new window on molecular imaging at a low dose of illumination. The ability to encode and decode polarized light pixel by pixel, coupled with the computational strength provided by artificial intelligence, is the running perspective of label-free optical microscopy. More specifically, the information-rich content Mueller matrix microscopy through its 16 elements offers multimodal imaging, an original data set to be integrated with other advanced optical methods. This dilates the spectrum of possible and potential applications. Here, we explore the recent advances in basic and applied research towards technological applications tailored for specific questions in biophysics and biomedicine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
“You walk into a place, you see there could be a great picture, but something is missing.
If that something comes, it can be a good photo—if not, it's nothing.”
(Gianni Berengo Gardin, Italian photographer)
1 Introduction
The revolutionary step from optical microscopy to optical nanoscopy [1] allows the formation of super-resolved images of biological molecules providing a spatial resolution better than the one imposed by the wavelength of the utilized electromagnetic (EM) radiation [2] and making possible the exploration of the biological nanoworld by visible light [3]. This result is achieved by implementing the smart concepts analysed by Toraldo di Francia [4] and Luksosz [5], namely: adding information to the light detection process and reducing the field of view [6]. The coupling with fluorescence labelling [7] of the object to be visualized permits to spatially resolve details of intricate extended cellular substructures [8] in a quantitative way [9].
Recent technological advances in sensing photons [10] allowed to extend the capabilities in the temporal domain adding spectroscopic fluorescence features, including lifetime [11,12,13,14] and anisotropy [15,16,17,18]. However, fluorescence labelling, considering natural or artificial labels, has shown some limitations under the super-resolved fluorescence optical microscopy lens. Among them, single-molecule imaging poses a problem related to possible effects on crowding in terms of re-organization of important biological macromolecules like chromatin in the cell’s [19, 20] nucleus and in terms of access of specific biological sites [21], as also reported by means of a correlative approach [22]. So far, fluorescence maintains key advantages like the biochemical specificity of the labelling probe and the brightness of the signal against a dark or dim background [23, 24]. The ability of controlling the on–off switching of fluorescence opened the pathway towards the development of super-resolved methods [25,26,27]. In such an advanced scenario, where the most recent results indicate the possibility of locating point light sources with a localization precision at the Angstrom level and a spatial resolution of 1 nm at room temperature and atmospheric pressure [28], Plato’s Allegory of the Cavern, reported in the book 7th of “Republic” (514a–520a) [29], Fig. 1, brings us back to the condition given by the relationship between what we know and what we understand which is not at all obvious.
The light microscope image presents a snapshot that can only do with additional information to draw more detailed conclusions. When you watch the historical photo of the bottle passing between two great past cyclists, Fausto Coppi and Gino Bartali, you need more information to decipher what really happened, Fig. 2.
Fluorescence, on its side, offers specificity and sensitivity to environmental conditions, Fig. 3, in exchange for inevitable contamination of the "crime scene" in terms of electrical charge and mechanical effects due to changes in local molecular crowding [30].
This sensitivity to molecular order is the key element for applications in biophysics and biomedicine starting from the cellular, organ and tissue levels for revealing those architectural dynamics that, for example, in living cells are strictly linked to the structural basis of cell function. Cells provide structure and function for all living things, from microorganisms to humans and contain the biological machinery for making the proteins, chemicals, and signals responsible for everything that determine the fate of living systems. The relationship between structure and function is complex and delicate at the very same time [31, 32]. Now, the debate about the impact of labelling on molecular organization is still open. This topic is not the scope of this review; however it is interesting to consider the tunable size of the major families of fluorescent probes, as reported in Fig. 4.
Different probes for labelling cellular structures in comparison with a cross section through a microtubule and an adenovirus capsid displayed to scale, from Sahal et al. [33]
However, one of the reasons for fluorescent labelling lies in the light–matter interaction: at visible wavelengths most of the biological molecules are almost transparent [34]. A perfectly aligned optical system would produce a very poor or no contrast in transmission when visible light interacts with most of the biological macromolecules contained in the biological cell that has also the property of being comparatively thin.
Considering the complex refractive index
as a molecular signature of the biological macromolecules, the real refractive index, n′, is prevalent on the imaginary part. The former affects the speed of light, while the latter is related to the absorption coefficient. Its overall value scales as the water content is the most abundant molecule in cells, accounting for 70% or more of total cell mass [31, 32].
Notwithstanding this, the optical microscope can produce cellular and molecular images endowed with contrast also when there are no labelled molecules. For example, this can be done exploiting scattering, intrinsic fluorescence, nonlinear light–matter interaction mechanisms like multiphoton excitation microscopy [35], second/third harmonic generation (SHG/THG) [36, 37], quantitative phase contrast (QPI) [38], Brillouin microscopy [39], pump-probe microscopy [40] and many other optical methods [41]. In general, label-free approaches have the potential to become highly relevant for studying the structure and function relationships in biological systems without the need of a contrast agent and for the development of new instrumentation in biophysics, biomedicine, and material sciences [42].
Now, when light interacts with matter, its polarization condition is sensitive and can be altered by the molecular organization in materials and biological systems. For this reason, our main interest in the framework of label-free optical methods is related to the polarization properties of light [43]. As recently reported by Oldenbourg [44], “the polarized light microscope was Shinya Inoue’s favourite tool for discovery [45]”, with which the control and detection of polarization changes offer a view beyond what we can imagine. It is a matter of fact that numerous animals are differentially sensitive to the vector orientation of polarized light [46], Fig. 5. However, the “classical” polarized optical microscope evolved towards three-dimensional imaging of all possible polarization parameters, including linear and circular birefringence, and circular dichroism [47] using high numerical aperture objectives [48, 49].
Now, Mueller matrix [50] microscopy can be seen as a natural evolution of polarized light microscopy in a label-free context that moved forward an increased number of applications in biology, medicine, pathology, and other fields of basic and applied science [51, 52]. Technological and computational advances have an important role, since Mueller matrix microscopy has the power of producing large data set and at the very same time is endowed with a robust mathematical model to characterize the optical properties of a medium at a certain wavelength in terms of polarization dynamics detectable in terms of forward, angular, and backward scattering.
The state of polarization of light as EM is determined by the evolution of the electric field of the travelling wave at a fixed point in space [53]. It is interesting that the polarization state, point by point, can be represented in a simplified and meaningful way by certain intrinsic properties like the intensity, the degree of linear and circular polarization that can be analysed in terms of appropriate compositions of Jones and Stokes vectors for coherent and incoherent superpositions, respectively. Staring from the formalism and the instrumental architecture needed to interpret polarization data, we will touch the base with some key applications in biophysics and biomedicine that have a strong impact in molecular oncology, pathogen detection and developmental biology and lay the foundation of an effective utilization of the modern advances in artificial intelligence.
2 Mueller matrix
Polarization describes the vectorial properties of an electromagnetic (EM) field in terms of the direction of its oscillation in the three-dimensional space [54]. As a convention, this term usually describes the vibration direction of the electric field component of the EM wave.
Considering an EM monochromatic plane wave oscillating at the optical frequency \(\omega \) and propagating along \(\widehat{z},\) its evolution in time and space is described by the following relation:
where \(\widehat{x}, \widehat{y}\) are the unitary vectors directed defining the \(x-y\) plane orthogonal to the propagation direction \(z\). The overall field is composed of two parts oscillating in two orthogonal directions. The relation between the amplitudes (\({A}_{x}\), \({A}_{y}\)) and the phases \({(\delta }_{x}\), \({\delta }_{y})\) of these two components defines the kind of polarization state. The most generic state of polarized light is termed elliptical polarization state (Fig. 6).
In Fig. 6, the polarization state is sketched in terms of the geometrical parameters of the ellipse, and its “handedness”, that is, whether the rotation around the ellipse is clockwise or counterclockwise. For any polarization form, the ellipse is characterized by [53]:
-
– its azimuth α, ranging in value from 0 ≤ α ≤ π;
-
– b and a, the lengths of its semi-minor and semi-major axes, respectively, and positive values of ε signifying right-handedness;
-
– ellipticity, tan|ε|= b/a, where -π/4 ≤ ε ≤ + π/4;
-
– diagonal angle β ranging 0 ≤ β ≤ π/2.
Here we have:
The amplitude and phase differences determine the ratio between the ellipse axes and their orientation. In degenerated cases, if the amplitudes of the two components are equal and their phase shift is null, the light is defined as linearly polarized, whereas if their phase difference is π/2, the field is circularly polarized (Fig. 7).
The Jones and the Stokes–Mueller formalisms are the two main physical frameworks used to describe the state of polarized light and their interaction with a medium [55]. Here, we address these two formalisms including some examples.
2.1 Jones formalism
The Jones formalism [56] deals with the vectorial description of the complex electric field components. A Jones vector made of 2 × 1 complex elements is used to describe the state of polarization of an EM field using the following representation:
where \(\delta \) is the phase difference \({\delta }_{y}-{\delta }_{x}\). The relative values of these two pairs of quantities define the overall polarization states. The complete evolution in time of the two components describes an ellipse in the plane orthogonal to the propagation direction, namely the \(x-y\) plane. Any pair of orthogonal Jones vectors is a basis of the overall EM field, so it can describe any state of polarized light. The vectors composing the most common bases, namely H–V, D–A, and R–L, are reported below.

On the other hand, the response of a medium upon excitation with polarized light is modelled through a 2 × 2 matrix with complex elements. Below, some common examples of polarizing optical elements, described through the Jones formalism, are reported.
A linear polarizer (LP) is an object exhibiting strong linear dichroism, that is, the selective ability to attenuate a specific polarization component, while the other remains unaltered upon transmission or reflection. Similarly, a phase retarder (HWP, QWP) selectively introduces a phase shift that depends upon the relative orientation of its optical axis and the polarization components of the EM field. Such matrices describe the polarimetric response of the optical elements with a certain orientation. When these optics are rotated along their azimuthal angle, their response can be adapted by applying the proper rotation matrix to the original Jones matrix if the rotation angle is known. Similar reasoning can be done for the vector describing light polarization. The rotation matrix is defined as
and its effect on the rotation of the optical elements or on the polarization of a light beam is defined by:
A limitation of Jones formalism is that it deals with a purely deterministic state of polarization (i.e. not depolarized). It is therefore an effective formalism to describe optical devices such as metamaterials and engineered materials in the field of material science, but is not the key approach to describe biological systems such as cells because of the complex optical properties and high degree of depolarization present therein [57].
2.2 Stokes–Mueller formalism
The Stokes–Mueller formalism deals directly with the intensity of polarization components of light (Fig. 8).
However, both the elements of the Stokes vector, used to describe the light polarization, and those of the Mueller matrix, used to model the medium response, are represented by real value coefficients. The elements of a generic Stokes vector express the intensity difference between orthogonally polarized components, as shown in the following definition:
where \({I}_{0}\) is the total intensity of the collected light and \({I}_{j}\) is the intensity of the jth polarization component with j = H, V, D, A, R, L, where H, V, D, R, L represent linear horizontal/vertical, + 45°/ − 45° and circular right/left polarization states, respectively. Below are reported the Stokes vectors for the most common states of fully polarized light.
Conversely to the Jones formalism, the Stokes vectors also allow the representation of partially polarized light. The polarization description is done by splitting the fully polarized and the unpolarized components as the sum of two vectors, as reported in the following relation:
Here, \(\gamma \) is the degree of polarization (DOP) of the light and it is defined as
whereas the coefficient \((1-\gamma )\) represents the amount of unpolarized light.
For a generic Stokes vector with physical realizability, the following relationship between its elements is always bounded by the following inequality:
where the two sides are equal in the case of completely polarized light (\(\gamma =1)\).
Finally, the Mueller matrix of an object interacting with a beam of polarized light has 16 real-valued coefficients, mi,j, linked by the Stokes vectors as
that are able to express its anisotropy properties, such as dichroism and birefringence.
The “object” is a combination given by the chain of k elements along the optical pathway, including the samples that are linearly connected as multiplication of the related Mueller matrix, known and unknown:
The following Mueller matrices are associated with some of the most common polarizing optical elements with the additional meaning that they can be employed to validate the performances of the related optical instrumentation.
The rotation matrix for the Stokes–Mueller formalism is:
and its effect on the rotation of the azimuthal angle of the optical elements or on the polarization of a light beam is described as follows:
Here, we report the extended results for the case of a linear polarizer and a half-wave plate that have been used as reference test samples. Their response can be proven by varying the azimuthal angles:
2.3 Mueller matrix information content
There are three main physical interactions that can cause changes of the polarization state of light, namely:
-
(1)
dichroism, associated with linear and circular as amplitude effects;
-
(2)
birefringence, associated with linear and circular phase effects;
-
(3)
depolarization, associated with spatial, temporal and/or spectral averaging effects.
Let us consider here diattenuators, retarders, and depolarizers as three basic optical elements that represent these effects considering the related Mueller matrices. This section will describe these elements and their associated Mueller matrices under the assumption that the optical waves are homogeneous and have orthogonal eigenstates of polarization [59].
More generally, any heterogeneous medium for the optical wave will generate depolarization [60]. Scattering is a process by which light is redirected in different directions by small particles or heterogeneous media, and it is an example of this phenomenon. When light undergoes scattering, it interacts with the particles, leading to a change in the polarization state of light. As light scatters in different directions, its polarization state can change randomly, resulting in a loss of the original polarization information.
Scattering can be influenced by several factors, including the size and shape of the scattering particles, the wavelength of light, and the concentration of particles in the medium. As a result, we will see, in another paragraph, how microscopy can utilize scattering to gain useful insights into a sample. An example of interest towards applications in biophysics and biomedicine is given by circular intensity differential scattering (CIDS), a technique that enables the analysis of a sample’s size, shape, and composition at the molecular and compaction level [61].
2.3.1 Diattenuators
Dichroism is defined as a difference in transmission (or reflection) between two orthogonal polarization states. A diattenuator device is a dichroic element that has an absorption anisotropy, and the intensity of the emerging beam depends on the polarization state of the incident wave. Scalar attenuation can be defined as
The quantification of the diattenuator depends on D values, where \(0\le D\le 1\) and \({T}_{\mathrm{max}}\), \({T}_{\mathrm{min}}\) are, respectively, the maximum and minimum energy transmittances. The energy transmittance for an unpolarized wave can be written as
In case D = 0, the transmittance or reflectance in intensity of this element does not depend on the state of polarization. When 0 < D < 1, the diattenuator is partial and \(D\) = 1 corresponds to a perfect polarizer. An attenuator element can polarize the incident wave linearly (linear dichroism), circularly (circular dichroism) or elliptically, but the most general case is that of a partial elliptical polarizer. Where the diattenuation of each component is dependent on the incident wave’s polarization, we define the diattenuation vector \(\overrightarrow{D}\) by
where \({D}_{\mathrm{H}}\) is the horizontal linear diattenuation and − 1 \(\le {\mathrm{D}}_{\mathrm{H}}\le \) 1; \({D}_{{45}^{\mathrm{o}}}\) is the linear diattenuation at + 45° with − 1 \(\le {D}_{{45}^{\mathrm{o}}}\le \) 1; \({D}_{\mathrm{C}}\) is the circular diattenuation, with − 1 \(\le {D}_{\mathrm{C}}\le \) 1. A diattenuator is linear if it does not present any circular diattenuation that means \({D}_{\mathrm{C}}=0\).
We can also write \(\overrightarrow{D}\) with the components of azimuth \({\mathrm{\alpha }}_{\mathrm{D}}\) and ellipticity \({\upvarepsilon }_{\mathrm{D}}\) of the state corresponding to the maximum transmittance, as follows:
Thus, a diattenuator is totally characterized by \(\overrightarrow{D}\) and \({T}_{0}\) through a matrix form [62,63,64].
where \(\left[{m}_{D}\right]\) is the reduced 3 × 3 diattenuation matrix written as
\(\widehat{D}\) is the unity vector representing the direction of the dichroic axis and \(\left[{I}_{3}\right]\) is the 3 × 3 identity matrix. An ideal linear polarizer is a well-known diattenuator that transmits light uniformly vibrating in a single plane while fully absorbing the orthogonal plane.
2.3.2 Retarders
A retarding element is also known as birefringent or as phase shifter, since it modifies the phase of E without altering its amplitude. It can be for example a uniaxial medium which has two different real refractive indices of \({n}_{1}\) and \({n}_{2}\ne {n}_{1}\). This leads to generate a phase delay between the two eigenstates associated with these two indices. We characterize a birefringent element of thickness \(l\) by its global delay or phase shift:
where \(0^\circ \le R \le 180^\circ \), \({\upphi }_{2}\) and \({\upphi }_{1}\) are the phase shifts associated with the orthogonal eigenstates of the birefringent, and \(\Delta n=\left|{n}_{2}-{n}_{1}\right|\) is the birefringence of the medium. This relation is valid for birefringent elements whose optical axis is perpendicular to the direction of wave propagation. Birefringent elements that can induce a delay by applying an electric field, such as liquid crystals, Pockels cells, or a mechanical constraint (e.g. photoelastic modulators), are essential in polarimetry to switch quickly between different states of polarization. Additionally, some biological samples, such as starch granules, exhibit linear birefringence, as demonstrated by the applications of polarization-resolved microscopy. As an example, a linear birefringent wave plate behaviour is sketched in Fig. 9.
Action of a birefringent wave plate, whose fast axis is oriented at 45°, on a light that is linearly polarized. The linearly polarized light enters the plate. The electric field can be resolved into two waves, parallel and perpendicular to the fast axis. In the plate, the perpendicular wave propagates slightly slower than the parallel one, resulting in an arbitrary phase shift between them to make elliptical polarization in general case at the output
2.3.3 Depolarizers
Contrary to the previous optical elements, a depolarizer transforms a totally polarized light to a partially polarized state-induced medium of the depolarization. This phenomenon appears as soon as a spatial, temporal, or spectral averaging of the polarimetric properties takes place at the detection level. The depolarization engendered by a medium essentially comes from the phenomenon of light scattering and strongly depends on the detection geometry used during the measurement. A depolarizing medium can be modelled in the Stokes–Mueller formalism, by a depolarizer, which in general form is the following:
where \(\left[{m}_{\Delta }\right]\) is the 3 × 3 reduced matrix. The simplest case is that of a total depolarizer, where any Stokes vector is transformed into another Stokes vector describing a totally unpolarized light. The associated Mueller matrix is then
If the induced depolarization is partial and the medium depolarization behaves in the same way to all the incident polarization states—isotropic depolarization—the Mueller matrix results in:
where \(0\le a\le 1\).
If the depolarization is different for each type of incident polarization state—depolarization anisotropy—the associated matrix turns into
where \(0\le (|a|, |b|, |\mathrm{c}|)\le 1\). To quantify this phenomenon, we use the average depolarization factor which corresponds to the mean of the principal factors of \(\left[{m}_{\Delta }\right]:\)
There are three cases when \(\Delta =0:\) the element does not depolarize, \(0<\Delta <1\); the element is a partial depolarizer and \(\Delta =1\),;the element is a total depolarizer. The phenomenon of depolarization can be generated after interaction of light with natural or manufactured elements such as milk, white paper or metal [65].
3 The instrument
Polarization-based imaging approaches have been used for many experimental applications and for the development of a range of practical devices due to their sensitivity to the medium structure and orientation. Figure 10 shows a very simple apparatus used to perform early polarimetric measurements at different angles by moving the light detector around a cuvette containing a sample suspended in solution.
A classical and simple setup for Mueller matrix polarimetry, from left to right: a photomultiplier mounted on a rotating arm endowed with a polarization state analyser for detection, a cuvette containing the sample, a photoelastic modulator as polarization state generator, and a laser source as illumination
This comparatively simple se-up is based on the general scheme reported in Fig. 11.
A simple conceptual and experimental design for polarization measurements [66]
The key question “Is light from the moon polarized?” was extended to a range of scientific questions. The growing interest for polarimetry pushed for a comparatively fast instrumental evolution implementing a simple and effective design as the one reported in Fig. 11. It is worth noting the broad spectrum of scientific studies based on polarization measurements from astrophysics [67, 68] to remote target detection [69, 70], from micro/nanoparticles in a turbid or highly scattering medium [71, 72] to oceanography [73].
Figure 12 reports the example of an early Mueller matrix polarimeter [74] designed and realized with the specific aim of studying high-order biopolymer organization in solution following the polarimetric setup of Fig. 11. Polarized light is collected and analysed at different angles exploiting the differential scattering originated by modulated circularly right and left illumination. A 632.8 nm light source, produced by a 5 mW He–Ne laser, passes through a Glan–Thompson polarizing prism. The beam, linearly polarized at 45″ to the scattering plane, impinges a photoelastic modulator (PEM) that changes alternatively its polarization with left and right handedness at the operating frequency of 50 kHz. The PEM exiting light illuminates the sample contained in a 25 mm diameter cylindrical Heralux quartz cuvette with a wall thickness of 2 mm and centred on the fixed part of a “homemade” rotary stage. The sample scatters light over all angles. Due to the comparatively large dimensions of the cuvette, the quartz entrance and exit windows can be seen as planar. The scattered radiation is collected in the scattering plane by a photomultiplier tube, calibrated over a large dynamic range, placed on the arm of the rotary stage, on which are mounted two apertures and a Glan–Thompson polarizer. The apertures restrict the size of the scattering volume captured by the PMT. The optical resolution of the instrument, in terms of acceptance angle, is 2″. The output current of the PMT enters a transimpedance preamplifier. The ac part of the signal is coupled to a lock-in amplifier, while the dc component is switched to be directly computer acquired by an A/D–D/A interface card. In this case, the setting time for each channel, in acquisition, is a maximum of 20 ps; the conversion time does not exceed 35 ps, and the throughput to memory is guaranteed for a minimum of 15,000 conversions/s. The lock-in amplifier, set to a time constant t = 100 ms (12 dB/oct), receives as signal reference the 50 kHz PEM controller “clock.” Demodulation occurs at frequencies F (50 kHz) and 2F (100 kHz). Changes in the PSG and PSA and selection of modulation frequencies allow to reconstruct several Mueller matrix elements [74].
It is worth noting that the main interest, in terms of applications, is focused on biophysics and biomedicine that are important pieces in the large “puzzle” of practical and potential developments towards problem-oriented instrumentation [51, 75]. Nowadays, Mueller matrix polarimetry is one of the most emerging, comprehensive, and effective method for the charming characteristic of providing the full polarimetric fingerprint of a specimen through its measurable 16 elements. Various methods have been developed, based on either temporal, spatial, or spectral polarization coding and decoding, to determine the elements, mij, of the Mueller matrix. Figure 13 shows the different combinations of the input and output signals that allow to extract combined values of the Mueller matrix elements.
Figure 14 reflects polarization changes due to the structural properties of the sample in the collection of the 16 images that can be formed by the Mueller matrix microscope [77].
Mueller matrix signature of Cetonia Aurata Beetle reporting, for example, about structural chirality in the element m03, also corresponding to m14 [78]
3.1 The Mueller matrix microscope
Now, considering optical microscopy, two approaches can be examined towards the measurement of the complete Mueller matrix elements mij across a sample in two dimensions (x, y). The former implements a full-field imaging approach using a digital camera endowed with full-field polarization coding and decoding stages [79]. The latter consists of a point-by-point laser scanning of the sample [80] that can benefit from the most promising optical microscopy approach based on image scanning microscopy coupled with a single-photon detector array used for advanced fluorescence microscopy applications [10, 81].
Regarding the first approach, it consists in using dynamic polarization optics [82, 83] for a sequential acquisition of at least 16 polarization-resolved intensity images of the sample attainable in few seconds up to few minutes. A snapshot Mueller matrix microscope, based on the simultaneous coding/decoding of the polarization states physically split in the sensor plane, is also used for diagnostic biomedical applications [79]. Polarization is encoded (0°, 45°, 90° and 135°) pixel by pixel, 4 by 4 to form a “superpixel”, enabling the reconstruction of the four Stokes coefficients in real time [84]. Using this solution, the main limitation is given by pixel cross talk and reduced spatial resolution [85]. The point-by-point scanning interrogation of the specimen allows for preserving the volumetric resolution by collecting the angular fingerprint. For this reason, this second solution is named polarization-resolved “scatterometry” [86].
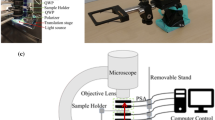
Now, we consider a modern confocal laser scanning microscope [87, 88] setup extended to Mueller matrix analysis by including a polarization state generator (PSG) and a polarization state analyser (PSA). For such a microscope, PSG and PSA optical modules are the core of the instrument, since they allow implementing the task of encoding and decoding the input–output polarization states. The polarization states generator encodes the polarization states from the incoming light source that can be emitted by a lamp, a laser diode, or a white laser source. This is clearly defined by the input Stokes vector, Sin, as described by Eq. (14). The combination of the four Sin states can also be described in the of the information content assigned to the “Degree of Linear and Circular Polarization”, DOLP and DOCP [89], respectively.
Since the impact of all the optical components in the illumination and detection pathways can be analysed component by component exploiting the superposition multiplicative effect derived by the utilization of the Mueller matrix formalism, the transformation of the polarized light after the sample is decoded by the polarization states analyser provides the output Stokes vector Sout collected by the detection unit. The combination of the PSA and the detector is also referred to as the polarization states detector (PSD). The turning point, in terms of information capacity provided by the Mueller matrix microscope, lies in the simple fact expressed by Eq. (15) that in an effective way describes how the polarization state of the input light changes upon interaction with the typically unknown sample.
So far, the main challenge for the implementation of this approach, considering the image contrast provided by each mi,j of each M, lies in an optical microscopy architecture optimized to adapt a proper methodology for encoding/decoding the polarization states in such a way as to retrieve pixel by pixel all the mi,j of the Mueller matrix representing the specimen. A decisive critical issue is given by the calibration of the full optical system. This must take into proper account the polarization state inaccuracy from the PSG and PSA errors combined with the polarimetric artefacts because of the multiple interactions with the optical features composing the optical components of the microscope.
Moreover, one has to consider that the specimen is interrogated by a diffraction limited spot usually driven by galvanometric mirrors, and that the light is transmitted or reflected by the in-focus illuminated volume element of the specimen before it is focused and collected by the detection unit. A general architecture is reported in Fig. 15.
Nowadays, the architecture based on the confocal microscope also implements the image scanning microscopy modality, using a SPAD (single-photon avalanche detector) array [nature methods con Castello] as an alternative to the classical PMT (photomultiplier tube) or hybrid single-point detection, which is a cross between a standard PMT and an avalanche photodiode [90]. As depicted in Fig. 15, the PSG is placed just before the objective lens, while the PSA is placed after the sample and the condenser in a transmission configuration. The SPAD array can be placed in an image plane as it happens for the pinhole. Its sensitive area is comparable to a typical pinhole size (one Airy disk size), and the module can be easily adapted to fit in any confocal microscope [88, 91, 92].
A key point in realizing and utilizing a Mueller matrix microscope is the accurate calibration of the whole architecture that measures the polarimetric contributions of the instrument components without sample: PSG, PSA, and the optical microscope fingerprints. This can be done using different strategies according to specific architectures and, more specifically, confocal modes [93]. A reference sample can be used to evaluate the robustness against noise propagation through the whole system. When using electro-optic devices like Pockels cells or photoelastic modulators [94], one can consider a global experimental noise propagation [95].
In confocal laser scanning mode, both pinhole and reflecting mirrors play an important role, and a recalibration of the system also when slight changes occur. However, the main assumption is that light is reflected by a “perfect mirror” at normal incidence, and that the fingerprint of the polarization features is independent of the direction of propagation of the light. Moreover, the microscope stand is usually composed of multiple optical elements that can change the polarization states defined by the PSG optics.
The strength of the Mueller matrix approach lies in the fact that one can isolate the Mueller matrix of the sample by using, component by component, matrices inversion:
where [Msample], [Mmicroscope] and [Mmes] are the Mueller matrices of the sample, the microscope and the total (microscope + sample), respectively.
Recently, full knowledge of the instrument performances at the pixel dwell time rate has been derived by a snapshot approach inspired by optical coherence tomography (OCT) [96].
The improvements obtained in terms of fast instrumental calibration has opened new windows towards the implementation of various imaging modalities on the very same scanning microscope [97].
3.1.1 Temporal domain encoding/decoding
The early instrumental developments in polarization-resolved microscopy were focused on the temporal control of the polarized and mainly realized by using rotating optical elements or electro-optics modulators. In single-point measurements, differential polarized intensity collection produces the determination of few Mueller matrix elements. Interestingly, such an approach allowed investigating and imaging of high-ordered macromolecules and biopolymers [98,99,100,101,102,103,104,105], including chromatin DNA [74, 106]. In biomedicine, Mueller matrix polarimetric imaging was specialized for blood cell characterization [107], reproduction and developmental studies [108, 109], and ophthalmology [110]. Without moving parts, the technique enabled the measurement of 16 successive double-pass images with an exposure time of 4 s, resulting in a full acquisition in 1 min. A further gain of using such an approach to study retinal tissues lies in the absence of damage. Figure 16 sketches the extraction of the Mueller matrix elements pixel by pixel, image by image.
Extraction of the MM(x,y) images from the measured intensities using a sequential approach. Here, t: temporal polarization states generation. IH, IV, I45, IRCP: intensities collected at the generated horizontal, vertical, 45° and right circular polarization (RCP) polarized states. Modified and adapted from [51]
In the confocal mode, an aperture slightly smaller in diameter than the Airy disc image is positioned in the image plane in front of the detector [91, 111, 112]. The strength of this method, since it was applied to spectroscopy studies in solution [113], lies in the ability of reducing out-of-focus blur signal coupled with an increased spatial resolution [114].
An early and notable application of confocal Mueller reflection microscope was for retinal diagnosis [93]. One has also to consider that the multiple reflection optics used as beamsplitters, Fresnel rhomb or Wollaston prism can be miniaturized thanks to the advance in designing and realizing metasurfaces [115,116,117]. More recently, a Mueller matrix polarimeter has been implemented into a nonlinear commercial transmission scanning microscope using simple motorized optical devices [118] allowing the acquisition of four images sequentially coded by four distinct polarization states (horizontal, vertical, 45° and right circular).
Another promising instrumental advance demonstrated, through highly scattering medium, depth-resolved imaging of cornea combining Mueller matrix elements in transmission and reflection configuration with nonlinear microscopy utilizing two-photon excitation fluorescence (TPEF) and second harmonic generation (SHG) [119]. In this case, both the PSG and PSAs are composed of a pair of liquid crystal variable retarders and a linear polarizer and the voltages are synchronized with the galvanometric scanners. The resulting images are built pixel by pixel from the measurements of four different PSA states and or each of one of the six PSG states, producing a dataset of 24 images. In this case, polarimetric contrast allowed to demonstrate that the random changes in the corneal model as a layered medium in the order of micrometre can strongly affect its polarization properties [119].
3.1.2 Spectral domain encoding/decoding
The previously reported examples are produced by acquiring a set of consecutive generated polarization-resolved images pointing out to some limitations towards in vivo microscopy imaging that can be overcome by using a series of multiple electro-optics devices such as photoelastic modulators (PEMs) [94, 120].
The analysis of the time variation of the detected signal is a channelled spectrum that can be represented in the Fourier domain by complex modulation amplitudes at different frequencies defined, for example, by the PEM operating frequencies. The photoelastic modulator is an optical device composed of a passive crystal subjected to periodic mechanical stress tuned on its resonant frequency that is material dependent. The passive crystal material, glass or quartz, influences both the operating frequency and the optical window transmission wavelengths. The functioning is based on the time-varying birefringence due to the photoelastic effect [94, 121]. A classical demodulation using PEMs is at 50 kHz for the first harmonic of the signal revealed by a lock-in-amplifier that allows using the second harmonic to select the detectable Mueller matrix elements.
To speed up the acquisition rate and the number of elements, the number of PEMs is increased combining the PSG and PSA elements [122]. The main advantage brought by adding multiple PEMs is that all the mij elements can be retrieved without any mechanically moving components.
This opens a new window for studying ultrafast conformational changes in biopolymers [123]. Considering the instrument, this means that the acquisition speed, 100 µs at 10 kHz repetition rate, is very close to the pixel dwell time used in confocal laser scanning microscopy [91, 124].
Fast polarization encoding evolved using passive elements such as birefringent plates [125]. In this case, the parallelization of polarization states in the spectral domain allows calculating the polarimetric response of a sample considering the single-channel spectrum I(ν), where ν is the optical frequency. Each channelled spectrum I(ν) is periodic and composed of discrete frequency integer multiples of the fundamental one. The fundamental frequency contains information about the thickness e and birefringence. One can potentially obtain fast scanning microscopy data set by optimizing the polarimeter stage’s optical elements [126].
The acquisition rate is limited by the spectrometer performances that can reach hundreds of MHz, and when operating in transmission allows multimodal imaging by including TPEF and SHG mechanisms of contrast [127].
Such a multimodal approach has been applied for label-free cancer cervix study by coupling Mueller matrix wide-field imaging with optical coherence tomography [128]. A cost to pay towards speed imaging lies in the key assumption that the sample is achromatic within the excitation wavelength range. Figure 17 shows a general optical scheme and the computational modalities associated with the spectral encoding/decoding Mueller matrix microscopy mode [129].
a Scheme of the architecture based on the spectral encoding/decoding of the polarization states. b Extraction of the mij images from the measured frequencies in the spectral approach. A human liver biopsy image in terms of retardance and orientation in the box as example (scale bar: 0.5 mm). PSG polarization states generator; PSA polarization states analyser; LP linear polarizer; RET retarder; Obj microscope objective; GS galvanometric scanner; TL tube lens; S sample; PMT photomultiplier tube (adapted from [129])
It is worth noting that the modelling of the instrument assumes that all the linear retarders used do not present any diattenuation or depolarization. Thus, a precise validation of the polarimetric properties of the sample must be achieved in the working spectral range.
3.2 A Zeeman effect laser-based microscope
The capability to achieve a fast encoding of polarization states to generate different Stokes vectors plays a crucial role in biological and medical applications and influences several critical aspects both on the experimental setup and in the imaging performances. Considering the architecture of a Mueller matrix microscope, as discussed before and shown in Fig. 15, the PSG produces pure states, which can be encoded spatially, temporally or spectrally. Using rotating birefringent optics, photoelastic modulators or Pockels cells for some applications can be comparatively slow. Moreover, suppose one wants to adapt a commercial confocal laser scanning microscope. In that case, there is the need for external devices, driving electronics and a power supply, and a careful alignment with the laser source to operate correctly.
In the 1980s from the Los Alamos National laboratories, there was an important improvement for the design of Mueller matrix microscopes and polarimeters with the advent of system, where the encoding of the polarization state is carried out by a Zeeman laser (ZL) that acts both as an illumination source and PSG [130].
This solution exploits the Zeeman effect [131] generated by an external magnetic field applied to the gain medium of the laser. Among the early implementations, it is worth noting the one related to differential phase measurements on scattered light from particles using a two-frequency Zeeman effect laser [132] emitting two frequencies of radiation 250 kHz apart. Furthermore, this illumination solution allowed excellent discrimination and reproducibility for various pure pollen and bacterial samples in suspension using polarization signature as shown in Fig. 18. The increased speed for encoding of such a solution also opened a perspective in flow cytometry applications [133].
The scattering of Zeeman laser light from pure aqueous suspensions of bacteria (adapted from [133])
3.2.1 The Zeeman effect laser as PSG
The first realization of a Zeeman laser is close to the invention of the He–Ne gas laser [134,135,136].
The Zeeman effect results in a peculiar light emission producing a dual-frequency, dual-polarization (DFDP) output [137]. Such a characteristic in the radiation emission is the core for utilizing the Zeeman laser as a light source and as PSG. As an example, a compact Zeeman laser was realized using an He–Ne laser tube mechanically coupled with seven arrays of rare-Earth magnets, arranged in a radial configuration, which can trigger the Zeeman effect in the gain medium. In this way, the laser output consists of a stable 632.8 nm beam made up of two laser lines spaced by 700 kHz. This design allows to obtain orthogonal circular polarization states and a beating signal can be generated for the emission of a stable 700 kHz spectral line. A stabilization unit made up of polarizing optics and fast photodiodes can be used to control the power of the two oscillating modes and as frequency counter of the optical beating. The use of a microcontroller allows using the photodiodes’ feedback signal to guarantee the laser stability and to minimize the effect of cavity length changes due to operating temperature variations [138].
This is an example of a fast PSG module that can be integrated in a confocal laser scanning architecture for live cell/tissue imaging.
3.2.2 Architecture of the microscope
The architecture of the laser scanning Mueller matrix microscope based on the Zeeman effect laser is shown in Fig. 19 [138].
Scheme of an optical scanning microscope using a Zeeman laser. The illumination travels through a scanning stage (SS), an expanding telescope (ET), followed by an excitation and detection objectives (OBJ) and the specimen (S). Following the interaction of the polarized illumination with the sample, the signal is split by a Wollaston prism (acting as PSA) and detected by two fast photodiodes (PDX and PDY). The inset shows the house built Zeeman effect laser. Modified after [138].
Here, to provide a practical example of an effective implementation are some details: a scanning stage (C2+ , Nikon Instruments, JA) is used for raster scanning of the laser beam. A 20×/0.5NA Nikon objective (DIC-M Plan Fluor, Nikon Instruments) is used to focus the polarized illumination on the specimen. The light after the interaction is collected by a 4×/0.13NA Nikon objective (CFI Plan Fluor, Nikon Instruments, JA). The polarization state is analysed using a Wollaston prism (WP10, Thorlabs, USA)—acting as PSA—aligned to separate the X-polarized and the Y-polarized components. These two output beams carry the interference signal of the Zeeman laser detected with a couple of Si-amplified photodiodes (PDX and PDY, PDA36A-EC, Thorlabs, USA). The photocurrents signals, split into two parts, are conditioned, digitalized and stored by the data acquisition system. The AC components are demodulated by a lock-in amplifier (LA, HF2LI, Zurich Instruments, CH) using an in-phase/quadrature (I/Q) demodulation. A phase reference is used to feed the LA with a reference beating signal coming from the stabilization unit of the Zeeman laser. Meanwhile, the DC components are directly obtained by the Nikon C2 workstation that also receives the LA output after demodulation. Nikon NIS-elements software is used for imaging. Analysis of the experimental data is performed employing custom Matlab (MathWorks, USA) routines.
3.2.3 Measurements of the Mueller coefficients
We use, considering the example provided in the previous paragraph, the intensity-based description of the polarization states carried out by the Stokes vector for two elements oscillating at the beating frequency (\({\omega }_{\mathrm{B} }= 2\pi {f}_{\mathrm{B}}\)) [139]:
Here, the polarization state of the light is encoded in time at high frequency. The experimental procedure needed to extract the values of the Muller matrix elements of the sample is done by analysing the amplitudes of the beating signal. All the optics in between these stages are neglected in the following mathematical description, since their capability to modify the polarization state is ideally null. The resulting Stokes vectors are obtained by the product of the Mueller matrices associated with the optical elements in the system, according to Eqs. (14), (31), and the input beam generated by the Zeeman laser, eq. zz1: so, the polarization state of light signals detected by the photodiodes are represented by
where the effect of the PSA is described by \([{M}_{X}]\) and \([{M}_{Y}]\) matrices, respectively, the Mueller matrices of a polarizer aligned along the X (0°) and Y (90°) axis. The specimen matrix is a general Mueller matrix \(\left[M\right]\).
The first element of each output Stokes vector is given by the intensity detected by the two photodiodes after the PSA:
Demodulating with the reference signal oscillating at \({\omega }_{B}\), the information about DC and AC components can be separated and simple calculations allow getting
With this configuration, six elements of the Mueller Matrix of the sample can be identified at the rate of the Zeeman beating frequency
The measured mij values are used to build pixel by pixel the corresponding six images.
Figure 20 shows the label-free imaging capabilities tested on starch granules embedded in potato cells: the m00 image shows the total transmitted intensity of the light and allows to see the clear shape and the outline of the granules. The m01 and m11 coefficients are related to birefringence and dichroism in the starch granules, demonstrating a specific molecular arrangement in the different regions of the starch granules.
Images of starch granules from a potato thin-cut slice obtained from the Zeeman laser method implemented into an optical scanning beam architecture. The pixel-by-pixel image is associated with the Mueller coefficients obtained by the analysis of the Zeeman beating signal. The pixel dwell time used for the acquisition was set equal to 10.8 μs, allowing full recordings of a 512 × 512 pixels image in less than 3 s with a scanning microscopy setup
4 Applications in biophysics and biomedicine
Questioning matter, the living, with visible light without using contrast mechanisms, has always been the challenge of the optical microscope. The challenge achieved with the phase contrast microscope by Zernike [140] opens an important window which, day after day, adds elements to the contrast mechanisms helpful in forming images through the microscope lens. Polarization, in the early elaborations of Stokes and Mueller, emerges among the “label-free” methods in applications in biophysics and biomedicine. Here, we selected some cases in a vast number of applications, and we report about a specific case study in molecular biology that is at the crossroads of biophysics and biomedicine [51, 141,142,143,144].
4.1 Selected applications
The Mueller matrix microscope has been demonstrated to be a fundamental apparatus for a wide range of biological applications, from biophysics to biomedicine. The core lies in the interpretation of the polarimetric response of the samples. Beyond being a label-free instrument, the strength can be related to the low dose of light needed and the number of different multimodal images, 16 as associated with the mij elements of the Mueller matrix. Indeed, biological matter exhibits more substantial depolarization than solid-state structures due to the random organization over all the illumination volume [145].
4.1.1 Ophthalmology
Mueller matrix laser scanning microscopy produces an approach for diagnosing a pathology at rate faster than the eye motion in tune with earlier methods proposed by optical coherence tomography imaging [128]. The potential advantages consist in the reduction of the exposure time that limits the damage to the eye. Moreover, the coupling with additional signal processing offers a large data set that can be examined by the images that can be formed starting from the mij intensities. The relationship between eye pathologies and the disorganization of thick tissues responsible of strong changes in the birefringence and depolarization parameters has been shown. Mueller matrix microscopy has the capability of quantifying these parameters. Moreover, an intelligent selection of the mij elements allows accelerating the process for encoding/decoding the polarization states [146, 147].
As strong points towards a Mueller matrix approach, one should consider the followings:
-
(1)
the ocular media and the retina in the human eye exhibit different polarization properties and techniques based on collecting the light scattered back into the retina in double pass must deal with several changes in polarization [148];
-
(2)
the fast motion and changes of the optical properties in the living eye require a high-speed rate encoding/decoding of the polarization states;
-
(3)
in terms of potential damage, the wavelength should be chosen wisely to reduce the radiation exposition and limit the absorption.
The influence of the eye pupil size on the birefringence was an important and early step towards applications in ophtamology [149]. For such an application, the numerical aperture of the objectives, determining the spatial resolution, could not be high for investigating at distance the specimen and for reducing strong polarization effects in the illumination direction. By means of the Mueller matrix microscopy approach, it is possible to quantify the polarimetric properties of the different compartments of the eye such as optic nerve, macula, cornea and retina, as shown in Fig. 21.
Quantitative imaging of the polarization parameters of the optical nerve head through the normalized average intensity m00; linear retardance—RL retardance orientation αR; depolarization index Dep; diattenuation D; diattenuation orientation. Adapted from [150]
However, as for other applications, it can be occasionally noted that the contrast distribution for some mij elements or derived parameters is completely blurred by the coupling effect of the back reflection with different scattering regions in the eye. Notwithstanding this, recent works have successfully overcome this issue by acquiring the elementary mij elements at 80 µm depth on extracted rat cornea using a confocal approach in both transmission and reflection [119]. This approach has gained an increased relevance in ophthalmology, since it can reveal how polarization heterogeneity in the polarization properties is related not only to the random distribution of the localized biological components, but is also influenced by other factors such as, for example, the glucose level [151].
4.1.2 Developmental biology and tissue organization
Since Mueller matrix microscopy offers the capability of tracking changes at distance in a non-invasive and label-free mode at low illumination power, many works have been focused on developing sample tailored solutions to investigate the modification of tissues induced by pathology at the cellular level [141, 152, 153]. This approach provides the chance of resolving sub-microscopic objects’ optical properties giving the capability of potentially quantifying pathologies at early stages.
The scoring of pathologies is performed by specialists and the accuracy of the analysis is subjective. This is the reason that the recent emerging methods proposed are evolving towards artificial intelligence solutions for data analysis. Here, one can measure physical changes due to oncological or neurological pathologies that result in detectable polarimetric effects comparatively to the healthy area. Thus, the Mueller matrix provides an interesting method for imaging confined localized structures and brings quantitative methods for staging the pathologies [51].
Among the variety of applications, Fig. 22 shows a detailed polarization fingerprint [109] at different developmental stages of the zebrafish (Danio rerio), a freshwater fish belonging to the minnow family (Cyprinidae) of the order Cypriniformes. This fish, native to South Asia, is an important and widely used vertebrate model organism in scientific research, for example in drug development towards pre-clinical developments [154]. However, for zebrafish, it has been proven than the imaging contrast from mapping the depolarization index comes from the thickness heterogeneity, while the retardance values are erased from the tissues and muscles.
Mueller matrix imaging of polarimetric parameters from fixed zebrafish embryos and larvae at 4 hpf (hours post-fertilization), 24 hpf, 48 hpf and 72 hpf. The images (a), (c), (e), (g) correspond to the total collected intensity element of the Mueller matrix, which is the m00 element. The images (b), (d), (f), (h) map diattenuation (D), retardance ® and its azimuthal orientations αD and αR, respectively. Adapted under a Creative Commons license [109]
So far, the Mueller matrix signature can provide a valued contribution to the understanding of the morphological changes of the animal during development within the structure and function relationships.
4.2 The case of circular intensity differential scattering (CIDS)
Circular dichroism (CD) is a powerful and well-established approach in the study of conformational properties of biomolecules [155, 156] that involves circularly polarized light, i.e. the differential absorption of left- and right-handed light [157,158,159]. This phenomenon is exhibited in the absorption bands of optically active chiral molecules. Circular dichroism is widely used in biophysics, biochemistry, structural biology and pharmaceutical chemistry [160]. The intensities of the circular left and right polarization of the light transmitted or scattered from chirally organized samples point to monitor structural similarities that are not identical to their mirrored image as function of the handedness of the molecule [161]. CD, which becomes CIDS when related to scattering (circular intensity differential scattering), corresponds to the m03 (also referred to as S14) element of the 4 × 4 scattering Mueller matrix. It has been proven that the scattering component, collected outside the absorption bands of the specimen, carries structural information at the single molecule level from long-range chiral structural motifs on a scale down to 1/10th–1/20th of the excitation wavelength, leading to information related to the structure and orientation of biopolymers in situ at the nanoscale [162, 163].
So far, CIDS has demonstrated, theoretically and experimentally, its sensitivity to (1) the characteristics of the chirality, such as their radius and pitch, (2) the handedness (left or right) of the molecules, and (3) the compaction of the chiral groups [164, 165].
However, CIDS emission is angularly defined as:
where IL and IR are, respectively, the detected intensities for the left and right circular polarization states and θ is the scattering angle [166].
CIDS is a label-free method for addressing questions related to DNA, RNA and protein organization like the delicate and intricate relationship between structure and function in chromatin, a fundamental biopolymer in the life and fate of any biological cell [167, 168].
The experimental procedure to measure CIDS is based on the Stokes–Mueller formalism. It is based on the generation and analysis of the multiple equations of polarization states coming from the interaction between the polarized light and the sample [76]. For typical CIDS measurements, the generation of the circular right and left polarization states is usually performed using electro-optic devices through a birefringent crystal stressed periodically using electro-optic devices [94] or, as recently reported, modulated laser sources like the Zeeman effect laser [138] combined with a lock-in detection at the reference frequency [74, 169].
Moreover, by adding multiple optical modulator devices in the setup, it is possible to achieve fast acquisition times for detecting temporal changes in the biopolymers’ organization [170]. Figure 23 sketches the different optical microscopy configurations developed for imaging CIDS pixel by pixel, inspired by early architecture: (a) a wide-field configuration using two Pockels cells [171]; (b) a solution using one photo elastic modulator synchronized with a lock-in amplifier and an XY translating sample holder [102, 172]; (c) a scanning microscope using a photoelastic modulator synchronized with a lock-in amplifier [143, 173].
Sketches of different architectures producing pixel-by-pixel CIDS imaging. PC Pockels cell; PEM photoelastic modulator; F monochromatic filter; LP linear polarizer; LA lock-in amplifier; PMT photomultiplier tube; GS galvanometric scanner. After [174]
The most relevant application of CIDS signature is the single-point measurement, in which the scattering from the whole illuminated sample volume is measured either as a function of the wavelength [175] or the scattering angle [176].
Among the increasing number of applications related to CIDS imaging [174], an important implementation regards its use for multimodal imaging of chromatin organization in cell nuclei [177]. Isolated HEK cell nuclei, also fluorescently labelled with a DNA binding fluorophore, were observed with both fluorescence and CIDS modality. This allowed to prove the validity of CIDS as an imaging mechanism for chromatin DNA compaction, as shown in Fig. 24 [177].
CIDS has been also proven to be helpful for virus and bacteria detection [178,179,180].
More recently, a computational approach reported about the potential of using chiral heterogeneity to detect SARS-CoV2 agents in water droplets [181].
5 A microscope in the computer machine
Today, optical microscopy has increased its role in science demonstrating an incredible capacity in producing large four-dimensional (x, y, z, t) data sets originated from biological systems [182] from molecules to cells, tissues and organs [183] by light interrogation boosted by unprecedented performances in terms of spatial and temporal resolution [28, 184] down to the nano- and picoscale [185], respectively. Such a data set can be considered the core for developing an artificial microscope aiming to transform a label-free interrogation of the sample into a molecular-rich fluorescence-based image [147]. The emerging role of the Mueller matrix approach is self-evident considering the 16 different simultaneous images that can be added to the “classical” multimodal collection. Figure 25 shows the hardware development of a multimodal architecture on the optical table. This is a crucial step towards the implementation and optimization of a microscope able to integrate different image formation abilities and taking advantage of a shared utilization of illumination sources and detection devices. A further component of the modern microscope that Italo Calvino considered in the “Lightness” chapter of the “Six Memos for the next Millennium” as the counterpart of the weight of the hardware is the software. This thread connects data stemming from the different light–matter interactions used: “Then we have computer science. It is true that software cannot exercise its powers of lightness except through the weight of hardware. But it is software that gives the orders, acting on the outside world and on machines that exist only as functions of software and evolve so that they can work out ever more complex programs. The second industrial revolution, unlike the first, does not present us with such crushing images as rolling mills and molten steel, but with “bits” in a flow of information traveling along circuits in the form of electronic impulses. The iron machines still exist, but they obey the orders of weightless bits.” [186].
The optical table arena where the multimodal microscope is under development to implement the liquid tunable microscopy concept @DiasproLab [187]
Figure 26 shows the overall architecture of a multimodal optical microscope using image correlation sensing. MOMIX is the acronym that includes the Mueller matrix microscope as an example of “liquid tunable microscopy” [187, 188]
MOMIX becomes intelligent when integrated with the “computer machine” taking advantage of the incredibly fast and impressive advances in the artificial and computational intelligence domains [189, 190].
However, there is a large interest for application in biophysics and medicine of artificial intelligence-based approaches to optical images, especially for the relevant impact they can have in decisional tasks in medicine, see as example Fig. 27 [191].
Example of machine learning workflow. Image patches are extracted from RGB liver photographs acquired during liver procurement. From each patch, textural and intensity features are extracted and added to clinical, biological, and radiological donor features. The features extracted from the training patches are used to train a semi-supervised SVM-SIL model (green boxes). The approach is semi-supervised because the ground-truth label is assigned to the whole image and not to the single patch [191]
So far, the design of a powerful intelligent AI-guided multimodal microscope is made by a computational core based on three modules: a convolutional neural network (CNN), a tensor-independent component analysis (tICA), an unsupervised machine learning technique able to extract relevant parameters and finally a supervised deep learning strategy to predict the cell fate. The ambitious target is to create a robust virtual environment to see “what we could not perceive before” with the potential of substituting biopsies by a label-free driven molecular image beyond the current state of the art [192]. Here, we show some preliminary developments related to the visualization of chromatin organization by predicting a fluorescence image originated by optical phas-dependent data [193].
5.1 The intelligent microscope: an example
The first module of the AI architecture for the intelligent microscope links the aim of predicting a fluorescence image from a label-free image by learning the specificity given by the presence of fluorescent proteins with the consequent result of facilitating the acquisition of a multimodal dataset, since multiple fluorescence images can be predicted and constructed from a single label-free image without the need to label the sample and deal with photobleaching and photodamage issues. The architecture is made up of a convolutional neural network in which the input is a label-free image, while the output is a single or multiple fluorescence image (Fig. 28).
Since the case study is related to chromatin compaction in the cell nucleus as function of potential cancer progression [194], the images for the training are HeLa cell nuclei fluorescently labelled with Hoechst 33,342 imaged by a confocal microscope (Nikon A1r MP, Nikon Instruments, Yokohama, Japan) also used to collect differential interference contrast (DIC) images as label-free components. Training was performed over 100 epochs, on 1700 images, with the MSE as a loss [185, 189, 195]. An example of the prediction using as input a DIC image [124] is shown in Fig. 29.
Example of prediction, the pixel size is 76.5 nm. On the left is the input label-free image, in the centre is the true fluorescence image, and on the right is the predicted image, which represent the output of the network [147]
In general, the multimodal microscope produces a large amount of data from the different light–sample interactions that need to be structured. Therefore, the second module aims to merge data coming from different mechanisms of contrast to automatically find patterns of related changes and extract common features across multiple modalities. Mathematically, the problem of representation is finding a projection of the data distribution that leads to some sort of ‘intrinsic’ coordinate system where the data structure is most apparent. To infer the same spatial patterns across modalities and therefore to fuse information, a tensor independent component analysis (tICA) approach appears adequate towards finding meaningful, spatially independent components in an unsupervised setting [147, 196].
So far, the multimodal image data set is modelled as a sum of components (Fig. 30), each of which can be expressed as the tensor product of one spatial map (\(n=1,\dots ,\mathrm{voxel}\)), one subject course (\(r=1, \dots , R\)) and one modality course (\(t=1, \dots , T\)).
where \({Y}_{n,t,r}\) is the dataset, i.e. a list of T different cells, imaged with T different contrast mechanisms, acquired using N voxel; \({X}_{n,i}\) are the spatial maps on n voxel for component i, where each component i has a single spatial map for all modalities; \({W}_{t,i}\) are the modality weightings for component i in modality t and tells which modality t uses to look into a specific component (and so into a spatial maps); \({H}_{i,r}\) are the weights for component i in subject r, form a link between the different modalities, and it is the appropriate place to look to find out which modalities drive a particular component; \({\varepsilon }_{n,t,r}\) is the noise, i.e. everything cannot be explained with the previous decomposition.
The last step is realized for leveraging those parameters obtained from tICA with the purpose of predicting in a supervised manner whether a cell, imaged through various different contrast methods, is pathological or normal. The use of Mueller matrix label-free data, able to generate after the first step molecular-rich images without the need of using contrast agents, has the property of predicting the fate of biological systems in time. Such a development has a tremendous potential impact on the real-time evaluation of a disease progression, on the decisional process during a surgical operation or on real-time evaluation of the effects of a pharmacological treatment [197]. A further challenge is provided by the hardware developments towards high-speed rate and the spectral information that give access to numerous measurable quantities at the same time. This advanced technique has been successfully proposed for real-time imaging through a polarization-resolved endoscopic approach that has the property of decoupling at any time the signatures of the sample from the optical fibre polarization transformation [198]. The perspective related to this marriage between artificial intelligence and label-free optical methods is given by the developments in the so-called “self-supervised” approaches that require no training data, apart from the input image itself [147, 199].
6 Conclusions
Optical label-free methods enabling the collection of signals from unlabelled biological molecules are on the way to providing microscopy images endowed with molecular specificity [200].
In parallel, advances in fluorescence microscopy provide data down to the Angstrom level [201]. Recent developments in fluorescent probes and acquisition strategies have resulted in spectacular growth in light microscopy in the life sciences, biophysics and biomedicine. The enormous benefit of these advances, coupled with artificial intelligence algorithms, is that they can provide molecular maps that can be linked to phase information, more in general label-free data. This fact is more than a perspective; it is today’s challenge for light microscopy. Basic and applied developments indicate new routes and significant chances towards practical aspects within the healthcare framework. The future without need for biopsies and the possibility of detecting in real time potential infectious contamination in shared spaces is the tip of an iceberg that will reveal something that we cannot see or imagine today. Although none of the label-free methods is able today to replace fluorescence microscopy or fulfil the needs of every label-free imaging experiment, it appears that we are rapidly moving to a turning point in label-free microscopy that holds great expectations for the future of these methods.
It is a matter of fact that today we can reveal hidden architectures (Fig. 31) and decipher what in the past we could not see without further explanations or hypothetical assumptions [202].
Polarization allows revealing what our eyes are not able to perceive; Antoine de Saint-Exupery brilliantly explained this in his novel “The Little Prince” in 1943. These drawings are a free adaptation of the related illustrations [202]
As in the early statements by Giuliano Toraldo di Francia, adding information increases our ability of mapping events beyond the physical limitations provided by the instruments in use [203].
It is an expanding field in terms of theoretical understanding and experimental applications with a focus on biomedicine. However, the developments using nonlinear light–matter interactions found an important perspective in structural molecular and biological imaging with the only drawback, in some cases, of requiring high illumination intensities [204]. Stokes polarimetry based on second harmonic generation provided new insights into collagen and skeletal muscle fibres [205] coupled with interesting solutions in terms of models and calibration procedures [206]. Moreover, approaches focused on optimizing imaging depth of scattering tissues [207] consolidated important label-free protocols towards the evaluation of bone-engineered materials [208]. Stokes–Mueller polarimetry is also bringing new information in those imaging processes dealing with multiphoton processes enabling the coupling of two/three photon imaging with third/second harmonic information content [209]. We can conclude that Mueller matrix microscopy is a powerful expanding field endowed with a high potential in biophysics and biomedicine.
Data availability
It is a review. However, data are available from the authors, if needed.
References
A. Diaspro, P. Bianchini, La Riv. Nuovo Cimento 43, 385–455 (2020). https://doi.org/10.1007/s40766-020-00008-1
E. Abbe, Arch. Mikrosk. Anat. 9, 413 (1873). https://doi.org/10.1007/BF02956173
A. Diaspro, Expedition into the Nanoworld An Exciting Voyage from Optical Microscopy to Nanoscopy (Springer Nature, Cham, 2022)
G.T. di Francia, Il Nuovo Cim. 9, 426 (1952). https://doi.org/10.1007/BF02903413
W. Lukosz, J. Opt. Soc. Am. 56, 1463 (1966)
C.J.R. Sheppard, Microsc. Res. Tech. 80, 590 (2017). https://doi.org/10.1002/jemt.22834
D.M. Jameson, Introduction to Fluorescence (CRC Press, Boca Raton, 2014)
A. Diaspro, Optical Fluorescence Microscopy (Springer, Berlin, 2011)
G. Weber, Annu. Rev. Biophys. Bioeng. 1, 553 (1972). https://doi.org/10.1146/annurev.bb.01.060172.003005
M. Buttafava, F. Villa, M. Castello, G. Tortarolo, E. Conca, M. Sanzaro, S. Piazza, P. Bianchini, A. Diaspro, F. Zappa, G. Vicidomini, A. Tosi, Optica 7, 755–765 (2020)
T.W.J. Gadella, T.M. Jovin, R.M. Clegg, Biophys. Chem. 48, 221–239 (1993)
G. Weber, J. Phys. Chem. 85, 949–953 (1981)
E. Gratton, S. Breusegem, J. Sutin, Q. Ruan, N. Barry, J. Biomed. Opt. 8, 381–390 (2003)
M. Castello, G. Tortarolo, M. Buttafava, T. Deguchi, F. Villa, S. Koho, L. Pesce, M. Oneto, S. Pelicci, L. Lanzanó, P. Bianchini, C.J.R. Sheppard, A. Diaspro, A. Tosi, G. Vicidomini, Nat. Methods 16, 175–178 (2019)
G.G. Stokes, Philos. Trans. 142, 463 (1852)
G. Weber, J. Chem. Phys. 55, 2399–2407 (1971). https://doi.org/10.1063/1.1676423
T. Parasassi, E.K. Krasnowska, L. Bagatolli, E. Gratton, J. Fluorescence 8, 365–373 (1998)
D.M. Jameson, J.A. Ross, Chem. Rev. 110, 2685–2708 (2010)
T. Misteli, Science 291(5505), 843 (2001)
K. Richter, M. Nessling, P. Lichter, J Cell Sci 120, 1673–1680 (2007)
M. Cosentino, C. Canale, P. Bianchini, A. Diaspro, Sci. Adv. 5, eaav8062 (2019)
S. Jadavi, P. Bianchini, O. Cavalleri, S. Dante, C. Canale, A. Diaspro, Microsc. Res. Tech. 84, 2472 (2021)
D.R. Sandison, W.W. Webb, Appl. Opt. 33, 603–615 (1994)
P.W. Hawkes, J.C.H. Spence, Springer Handbook of Microscopy (Springer Nature, Cham, 2019)
M. Heilemann, E. Margeat, R. Kasper, M. Sauer, P. Tinnefeld, JACS 127, 3801 (2005)
S.W. Hell, Science 316, 1153 (2007). https://doi.org/10.1126/science.1137395
S.W. Hell, S.J. Sahl, M. Bates, X. Zhuang, R. Heintzmann, M.J. Booth, J. Bewersdorf, G. Shtenge, H. Hess, P. Tinnefeld, A. Honigmann, S. Jakobs, I. Testa, L. Cognet, B. Lounis, H. Ewers, S.J. Davis, C. Eggeling, D. Klenerman, K.I. Willig, G. Vicidomini, M. Castello, A. Diaspro, T. Cordes, J. Phys. D Appl. Phys 48, 443001 (2015). https://doi.org/10.1088/0022-3727/48/44/443001
M. Weber, H. von der Emde, M. Leutenegger, P. Gunkel, S. Sambandan, T.A. Khan, J. Keller-Findeisen, V.C. Cordes, S.W. Hell, Nat. Biotechnol. 41, 569–576 (2023)
L. Rauch, Imagery and allegory in philosophy, in Allegory Revisited. Analecta Husserliana, vol. 41, ed. by A.T. Tymieniecka (Springer, Dordrecht, 1994)
S. Lecinski Sarah, J.W. Shepherd, K. Bunting, L. Dresser, S.D. Quinn, C. MacDonald, M.C. Leake, Interface Focus 12, 2022004 (2022)
B. Alberts, R. Heald, A. Johnson, D. Morgan, M. Raff, K. Roberts, P. Walter, Molecular Biology of the Cell, 7th edn. (W. W. Norton & Company, New York, 2022)
R. Milo, R. Phillips, Cell Biology by the Numbers (Garland Science, NY, 2015)
S. Sahl, S.W. Hell, S. Jakobs, Nat Rev Mol Cell Biol 18, 685–701 (2017)
I. Tinoco Jr., Methods Enzymol. 246, 13–18 (1995)
A. Diaspro, G. Chirico, M. Collini, Q. Rev. Biophys. 38, 97 (2005)
I. Freund, M. Deutsch, Opt. Lett. 11, 94–96 (1986)
Y. Barad, H. Eisenberg, M. Horowitz, Y. Silberberg, Appl. Phys. Lett. 70, 922–924 (1997)
Y. Park, C. Depeursinge, G. Popescu, Nat. Photon. 12, 578–589 (2018)
G. Scarcelli, S.H. Yun, Nat. Photon. 2, 39–43 (2008)
G. Zanini, K. Korobchevskaya, T. Deguchi, A. Diaspro, P. Bianchini, ACS Nano 13, 9673 (2019)
N. Mazumder, N.K. Balla, G.Y. Zhuo, Y.V. Kistenev, R. Kumar, F.J. Kao, S. Brasselet, V.V. Nikolaev, N.A. Krivova, Front. Phys. 7, 170 (2019)
V. Astratov, Label-Free Super-Resolution Microscopy (Springer, Cham, 2019)
W.A. Shurcliff, Polarized Light. Production and Use (Harvard University Press, Harvard, 1962)
R. Oldenbourg, Microsc. Microanal. 28(Suppl 1), 1550–1552 (2022)
S. Inoué, W.L. Hyde, J. Bioche. Biophys. Cytol. 3, 831–838 (1957)
A. Le Floch, G. Ropars, J. Enoch, V. Lakshminarayanan, Vis. Res. 50, 2048–2054 (2010)
R. Oldenbourg, Nature 381, 811–812 (1996)
R. Oldenbourg, P. Török, Appl. Opt. 39, 6325–6331 (2000)
C.J.R. Sheppard, M. Gu, J. Mod. Opt. 40, 1631–1651 (1993)
H. Mueller, J. Opt. Soc. Am. 38, 661–662 (1948)
A. Le Gratiet, A. Mohebi, F. Callegari, P. Bianchini, A. Diaspro, Appl. Sci. 11, 1632 (2021)
K.U. Spandana, K.K. Mahato, N. Mazumder, Lasers Med Sci. 34, 1283–1293 (2019)
J.J. Gil, R. Ossikovski, Polarized Light and the Mueller Matrix Approach (CRC Press Taylor & Francis, New York, 2016)
M. Born, E. Wolf, Principles of Optics: Electromagnetic Theory of Propagation, Interference and Diffraction of Light (Pergamon Press, Oxford, 1959)
S.N. Savenkov, Jones and Mueller matrices: structure, symmetry relations and information content, in Light Scattering Reviews, vol. 4, ed. by A.A. Kokhanovsky (Springer, Berlin, Heidelberg, 2019)
R.C. Jones, J. Opt. Soc. Am. 31, 488–493 (1941)
E. Kuntman, M.A. Kuntman, O. Arteaga, J. Opt. Soc. Am. A 34, 80–86 (2017)
J.J. Gil, Eur. Phys. J. Appl. Phys. 40, 1–47 (2007)
R.W. Wood, Physical Optics (Macmillan, New York, 1911)
V.V. Tuchin, Physics 40, 495 (1997)
F. Allen, C. Bustamante (eds.), Applications of Circularly Polarized Radiation Using Synchrotron and Ordinary Sources (Plenum Press, New York, London, 1984)
S.Y. Lu, R.A. Chipman, J. Opt. Soc. Am. A 13, 1106–1113 (1996)
C.J.R. Sheppard, A. Bendandi, A. Le Gratiet, A. Diaspro, Photonics 9, 394 (2022)
C.J.R. Sheppard, M. Castello, A. Diaspro, J. Opt. Soc. Am. A 33, 741–751 (2016)
F. Perrin, J. Chem. Phys. 10, 415 (1942)
W.S. Bickel, W.M. Bailey, Am. J. Phys. 53, 468–478 (1985)
R. Anche, A.K. Sen, G. Anupama, K. Sankarasubramanian, W. Skidmored, J. Astron. Telesc. Instrum. Syst. 4, 01800 (2018)
C. Patty, J. Kate, W. Jan Buma, R. van Spanning, G. Steinbach, F. Ariese, F. Snik, Astrobiology 19, 1221–1229 (2019)
J. Tyo, D. Goldstein, D. Chenault, J. Shaw, J. Appl. Opt. 45, 5453–5469 (2006)
J. Fade, S. Panigrahi, A. Carre, L. Frein, C. Hamel, F. Bretenaker, H. Ramachandran, M. Alouini, Appl. Opt. 53, 3854–3865 (2014)
A. Doronin, C. Macdonald, I. Meglinski, J. Biomed. Opt. 19, 025005 (2014)
V. Sankaran, J.T. Walsh, D.J. Maitland, J Biomed Opt. 7, 300 (2002)
K.J. Voss, E.S. Fry, Appl. Opt. 23, 4427–4439 (1984)
A. Diaspro, M. Bertolotto, L. Vergani, C. Nicolini, IEEE Trans. Biomed. Eng. 38, 670–678 (1991)
H. Honghui, L. Ran, Z. Nan, L. Pengcheng, C. Zhenhua, L. Xi, M. Hui, J. Lightwave Technol. 37, 2534–2548 (2019)
P.C. Thompson, S.R. Bottigerr, E.S. Fry, Appl. Opt. 19, 1323–1332 (1980)
Oriol Arteaga Lab, http://www.mmpolarimetry.com
O. Arteaga, M. Baldrís, J. Antó, A. Canillas, E. Pascual, E. Bertran, Appl. Opt. 53, 2236–2245 (2014)
J. Qi, C. He, D. Elson, Biomed. Opt. Expr. 8, 4933–4946 (2017)
C. Chen, X. Chen, Y. Shi, H. Gu, H. Jiang, S. Liu, Appl. Sci. 8, 2583 (2018)
C.B. Müller, J. Enderlein, Phys. Rev. Lett. 104, 198101 (2010)
K. Lai, Y. Yu, Q. Sui, D. Wang, Z. Li, Opt. Expr. 31, 12175–12188 (2023)
B. Koch, R. Noé, D. Sandel, V. Mirvoda, Opt. Expr. 22, 8259–8276 (2014)
P. Lapray, L. Gendre, A. Foulonneau, L. Bigué, Int. J. Circuit Theory Appl. 46, 1675–1689 (2018)
S. Powell, V. Gruev, Opt. Expr. 21, 21039–21055 (2013)
N.G. Orji, M. Badaroglu, B.M. Barnes et al., Nat. Electron. 1, 532–547 (2018)
A.D. Elliott, Curr. Protoc. Cytom. 92, e68 (2020)
G. Tortarolo, A. Zunino, F. Fersini et al., Nat. Commun. 13, 7723 (2022)
G. Stokes, Trans. Camb. Philos. Soc. 9, 339–416 (1992)
J. Jonkman et al., Nat. Protoc. 15, 1585–1611 (2020)
A. Diaspro (ed.), Confocal and Two-Photon Microscopy: Foundations, Applications and Advances (Wiley, New Jersey, 2001)
I. Nepita et al., Appl. Sci. 13, 1556 (2023)
D. Lara, C. Dainty, Appl. Opt. 45, 1917–1930 (2006)
J. Kemp, J. Opt. Soc. Am. 59, 950–954 (1969)
D. Sabatke et al., Opt. Lett. 25, 802–804 (2000)
B.E. Bouma et al., Nat. Rev. Methods Primers 2, 79 (2022)
C. Macias-Romero, P. Torok, J. Eur. Opt. Soc. 7, 12004 (2012)
A. Schneider, M.J. Rosenheck, Proc. Natl. Acad. Sci. USA 66, 793–798 (1970)
B. Dorman, M.F. Maestre, Proc. Natl. Acad. Sci. USA 70, 255–259 (1973)
I.J. Tinoco, M.F. Maestre, C. Bustamante, D. Keller, Pure Appl. Chem. 56, 1423–1428 (1984)
D. Keller, C. Bustamante, M. Maestre, I. Tinoco, Proc. Natl. Acad. Sci. USA 82, 401–405 (1985)
D. Beach, K. Wells, F. Husher, C. Bustamante, Rev. Sci. Instrum. 58, 1987–1995 (1987)
M. Kim, C. Keller, C. Bustamante, Biophys. J. 242, 313–356 (1987)
L. Finzi, L. Ulibarri, C. Bustamante, Differential polarization imaging. V. Numerical aperture effects and the contribution of preferential scattering and absorption to the circular dichroism images. Biophys. J. 59, 1183–1193 (1987)
V. Gupta, J. Kornfield, Polarization modulation laser scanning microscopy: a powerful tool to image molecular orientation and order. Rev. Sci. Instrum. 65, 2823–2828 (1994)
A. Le Gratiet, R. Marongiu, R. Diaspro, Circular intensity differential scattering for label-free chromatin characterization: a review for optical microscopy. Polymers 12, 2428 (2020)
W. Mickols, M. Maestre, I. Tinoco, S. Embury, Visualization of oriented hemoglobin S in individual erythrocytes by differential extinction of polarized light. Proc. Natl. Acad. Sci. USA 82, 6527–6531 (1985)
K. Wells, D. Beach, D. Keller, C. Bustamante, An analysis of circular intensity differential scattering measurements: Studies on the sperm cell of Eledone cirrhosa. Biopolymers 25, 2043–2064 (1986)
A. Le Gratiet, M. D’Amora, M. Duocastella, R. Marongiu, A. Bendandi, S. Giordani, P. Bianchini, A. Diaspro, Zebrafish structural development in Mueller-matrix scanning microscopy. Sci. Rep. 9, 1–10 (2019)
J.M. Bueno, P. Artal, Double-pass imaging polarimetry in the human eye. Opt. Lett. 24, 64–66 (1999)
C.J.R. Sheppard, A. Choudhury, Opt. Acta Int. J. Opt. 24, 1051–1073 (1977)
C.J.R. Sheppard, M.D. Sharma, J. Mod. Opt. 48, 1517–1525 (2001)
H. Naora, Science 114, 279 (1951)
C.J.R. Sheppard, Appl. Sci. 11, 8981 (2021)
Y. Intaravanne, X. Chen, Nanophotonics 9, 1003–1014 (2020)
C.J.R. Sheppard, A. Bendandi, A. Le Gratiet, A. Diaspro, Photonics 9, 88 (2022)
C.J.R. Sheppard, A. Bendandi, A. Le Gratiet, A. Diaspro, J. Opt. Soc. Am. A 35, 2058–2069 (2018)
N. Mazumder, L. Xiang, J. Qiu, F.J. Kao, Sci. Rep. 7, 1 (2017)
V. Le, I. Saytashev, S. Saha, P. Lopez, M. Laughrey, J. Ramella-Roman, Biomed. Opt. Expr. 11, 5982–5994 (2020)
S. Alali, A. Gribble, I.A. Vitkin, Opt. Lett. 41, 1038–1041 (2016)
A. Hunt, D.A. Huffman, Rev. Sci. Instrum. 44, 1753–1762 (1973)
O. Arteaga, J. Freudenthal, B. Wang, B. Kahr, Appl. Opt. 51, 6805–6817 (2012)
O. Arteaga, Z. El-Hachemi, R. Ossikovski, Opt. Expr. 27, 6746–6756 (2019)
J. Pawley, Handbook of Biological Confocal Microscopy (Springer, Boston, 2006)
K. Oka, T. Kaneko, Opt Expr. 11, 1510–1519 (2003)
M. Dubreuil, S. Rivet, B. Le Jeune, J. Cariou, Opt. Expr. 15, 13660–13668 (2007)
M. Dubreuil, F. Tissier, L. Le Roy, J.P. Pennec, S. Rivet, M.A. Giroux-Metges, Y. Le Grand, Biomed. Opt. Expr. 9, 6350–6358 (2018)
J. Chue-Sang, Y. Bay, S. Stoff, M. Gonzalez, N. Holness, J. Gomes, R. Jung, A. Gandjbakhche, V. Chernomordik, J. Ramella-Roman, J. Biomed. Opt. 22, 1–9 (2017)
A. Le Gratiet, M. Dubreuil, S. Rivet, Y. Le Grand, Opt. Lett. 41, 4336–4339 (2016)
R. Johnston, Interferometric Apparatus andMethod For Detection and Characterization of Particles Using Light Scattered Therefrom (Los Alamos National Laboratories, 1988). USA patent 4,764,013
P. Thalau, T. Ritz, H. Burda, R.E. Wegner, R. Wiltschko, J. R. Soc. Interface 3, 583–587 (2006)
R.G. Johnston et al., Appl. Opt. 25, 3566–3572 (1986)
R.G. Johnston, Proc. SPIE New Technol. Cytometry 1063, 198–204 (1989)
O. Svelto, Principles of Laser, 5th edn. (Springer, New York, 2010)
A. Javan, W.R. Bennett, D.R. Herriott, Phys. Rev. Lett. 6, 106–110 (1961)
R. Paananen, C.L. Tang, H. Statz, Proc. IEEE. 51, 63–69 (1963)
P. Zeeman, Nature 55, 347 (1897)
F. Callegari, A. Le Gratiet, A. Zunino, A. Mohebi, P. Bianchini, A. Diaspro, Front. Phys. 9, 758880 (2021)
W.M. McClain, W.H. Jeng, B. Pati, Y. Shi, D. Tian, Appl. Opt. 33, 1230 (1994)
F. Zernike, How I Discovered Phase Contrast Nobel Lecture. www.nobelprize.org/prizes/physics/1953/zernike/lecture/. Accessed December 11, 1953.
S. Alali, A. Vitkin, J. Biomed. Opt. 20, 61104 (2015)
J.C. Ramella-Roman, H. Ma, T. Novikova, D.S. Elson, I.A. Vitkin (eds.), Polarized Light and Optical Angular Momentum for Biomedical Diagnostics Proc of SPIE 12382 (SPIE, Bellingham, Washington, USA, 2023)
V. Gupta, J. Kornfield, Rev. Sci. Instrum. 65, 2823–2828 (1994)
D.N. Ignatenko, A.V. Shkirin, Y.P. Lobachevsky, S.V. Gudkov, Appl. Sci. 12, 5258 (2022)
V. Tuchin, L. Wang, D. Zinnyakov, Opt. Polar. Biomed. Appl. 4, 45–67 (2006)
T. Novikova, J.C. Ramella-Roman, Opt. Lett. 47, 5549–5552 (2022)
A. Diaspro, P. Bianchini, L. Cuneo, The artificial microscope, in Proc. of European Optical Society Annual Meeting, 2023.
J.M. Bueno, P. Artal, Opt. Lett. 24, 64–66 (1999)
J.M. Bueno, M.C.W. Campbell, Confocal scanning laser ophthalmoscopy improvement by use of Mueller-matrix polarimetry. Opt. Lett. 27, 830–832 (2007)
K. Twietmeyer, R. Chipman, A. Elsner, Y. Zhao, D. VanNasdale, Opt. Expr. 16, 21339–21354 (2008)
P. Westphal, J.M. Kaltenbach, K. Wicker, Biomed. Opt. Expr. 7, 1160–1174 (2016)
J. Qi, D. Elson, J. Biophoton. 10, 950–982 (2017)
H. He, R. Liao, N. Zeng, L. Pengcheng, Z. Chen, X. Liu, M. Hui, Light. Technol. 37, 2534–2548 (2019)
R.C. Van Wijk, E.H. Krekels, T. Hankemeier, H.P. Spaink, P.H. Van der Graaf, Drug Discov. Today Dis. Model. 22, 27–34 (2017)
K. Nakanishi, N. Berova, R. Woody, Circular Dichroism: Principles and Applications (VCH, New York, 1994)
L.D. Barron, Molecular Light Scattering and Optical Activity (Cambridge University Press, UK, 2004)
I. Tinoco Jr., Adv. Chem. Phys. 4, 113 (1962)
W.C. Johnson, I. Tinoco, Biopolymers 7, 727–749 (1969)
W.C. Johnson Jr., I. Tinoco Jr., Biopolymers 8, 715–731 (1969)
B.A. Wallace, Curr. Opin. Struct. Biol. 58, 191–196 (2019)
I. Tinoco, M. Mickols, M.F. Maestre, C. Bustamante, Annu. Rev. Phys. Chem. 16, 319–349 (1987)
C. Bustamante, Circular Intensity Differential Scattering of Chiral Molecules. Ph.D. thesis (Berkeley University, Berkeley, CA, USA, 1980)
C. Bustamante, I. Tinoco, M. Maestre, Proc. Natl. Acad. Sci. USA 80, 3568–3572 (1983)
M.W. Ashraf, A. Diaspro, Opt. Commun. 522, 128639 (2022)
M.W. Ashraf, A. Le Gratiet, A. Diaspro, Polymers 13, 3422 (2021)
A. Diaspro, C. Nicolini, Cell Biophys. 10, 45–60 (1987)
J. Widom, Annu. Rev. Biophys. Biomol. Struct. 27, 285–327 (1988)
K.E. van Holde, Chromatin (Springer-Verlag, New York, 1989)
J.E. Katz, S. Wells, D. Ussery, C. Bustamante, M.J. Maestre, Rev. Sci. Instrum. 55, 1574–1579 (1984)
O. Arteaga, J. Freudenthal, B. Wang, S. Nichols, B. Kahr, Chim. Oggi/Chem. Today 30, 6–9 (2012)
W. Mickols, C. Bustamante, M. Maestre, I. Tinoco, S. Embury, Nat. Biotechnol. 3, 711–714 (1985)
L. Finzi, C. Bustamante, G. Garab, C. Juand, Proc. Natl. Acad. Sci. USA 86, 8748–8752 (1986)
A. Le Gratiet, L. Pesce, M. Oneto, R. Marongiu, G. Zanini, P. Bianchini, A. Diaspro, OSA Contin. 1, 1068–1078 (2018)
A. Le Gratiet, R. Marongiu, A. Diaspro, Polymers 12, 2428 (2020)
S. Yoo, Q.H. Park, Sci. Rep. 5, 1–8 (2015)
M. Maestre, G. Salzman, R. Tobey, C. Bustamante, Biochemistry 24, 5152–5157 (1985)
R. Marongiu, A. Le Gratiet, L. Pesce, P. Bianchini, A. Diaspro, OSA Continuum 3, 1770–1780 (2020)
G.C. Salzman, C.T. Gregg, Biotechnology 2, 243–248 (1983)
G.C. Salzman, J.K. Griffith, C.T. Gregg, Appl. Environ. Microbiol. 44, 1081–1085 (1982)
A. Diaspro, G. Radicchi, C. Nicolini, IEEE Trans. Biomed. Eng. 42, 1038–1043 (1995)
M.W. Ashraf, R. Ranjan, A. Diaspro, J. Opt. Soc. Am. B 38, 1702–1709 (2021)
M. Hartley, G.J. Kleywegt, A. Patwardhan, U. Sarkans, J.R. Swedlow, A. Brazma, J. Mol. Biol. 434, 11 (2022)
L. Teodori, A. Crupi, A. Costa, A. Diaspro, S. Melzer, A. Tarnok, J. Biophoton. 10, 24–45 (2017)
S.C.M. Reinhardt, L.A. Masullo, I. Baudrexel et al., Nature 617, 711–716 (2023)
M.A. Boden, Artif. Intell. 103, 347–356 (1998)
I. Calvino, Six Memos for the Next Millennium. The Charles Eliot Norton Lectures 1985–1986 (Penguin Books, USA, 1988)
A. Diaspro, N. Anthony, P. Bianchini, I. Cainero, M. Di Bona, L. Lanzanò, A. Le Gratiet, R. Marongiu, M. Oneto, S. Pelicci, L. Pesce, Microsc. Microanal. 24, 1368 (2018)
R. Won, Nat. Photon. 12, 259–261 (2018)
L. von Chamier, R.F. Laine, R. Henriques, Biochem. Soc. Trans. 47, 1029–1040 (2019)
M. Asim, Y. Wang, K. Wang, P.Q. Huang, IEEE Trans. Emerg. Top. Comput. Intell. 4, 742–763 (2020)
M. Cesaretti, R. Brustia, C. Goumard, F. Cauchy, N. Poté, E. Dondero, C. Paugam-Burtz, F. Durand, V. Paradis, A. Diaspro, L. Mattos, O. Scatton, O. Soubrane, S. Moccia, Liver Transpl. 26, 1224–1232 (2020)
W. Chen, M. Tang, L. Wang, Sensors 23, 2882 (2023)
T. Cacace, V. Bianco, P. Ferraro, Opt. Lasers Eng. 135, 106188 (2020)
T.C.Q. Burgers, R. Vlijm, Chromosoma 132, 191–209 (2023)
A. Bisercic, M. Nikolic, M. van der Schaar, B. Delibasic, P. Lio, A. Petrovic, arXiv 2306.05052v1 (2023)
A.R. Groves, C.F. Beckmann, S.M. Smith, M.W. Woolrich, Neuroimage 54, 3 (2011)
H. Chen, C. Gomez, C.M. Huang et al., npj Digit. Med. 5, 156 (2022)
S. Rivet, A. Bradu, A. Podoleanu, Opt. Expr. 23, 23768–23786 (2015)
Y. He, J. Yao, L. Liu, Y. Gao, J. Yu, S. Ye, H. Li, W. Zheng, Photon. Res. 11, 1–11 (2023)
D. Evanko, Nat. Methods 7, 36 (2010)
A. Curd, Nature 617, 681–682 (2023)
A. de Saint-Exupéry, Le Petit Prince (Editions Gallimard, Paris, 2013)
R. Pratesi, L. Ronchi, C. Vasini (eds.), Waves, Information, and Foundations of Physics: A Tribute to Giuliano Toraldo Di Francia on His 80th Birthday, Florence, 15–17 September 1996 (Italian Physical Society, Editrice Compositori, Bologna, 1998)
S. Brasselet, Adv. Opt. Photon. 3, 205 (2011)
N. Mazumder, F.J. Kao, Lasers Med. Sci. 36, 1161–1167 (2021)
N. Mazumder, J. Qiu, M.R. Foreman, C.M. Romero, P. Török, F.J. Kao, Biomed. Opt. Expr. 4, 538–547 (2013)
S.Y. Chen, Z.T. Su, D.J. Lin, M.X. Lee, M.C. Chan, S. Das, Results Phys. 28, 104653 (2021)
C.W. Hung, N. Mazumder, D.J. Lin, W.L. Chen, S.T. Lin, M.C. Chan, G.Y. Zhuo, Microsc. Microanal. 27, 587–597 (2021)
X. Li, W. Liu, F. Goudail, S.C. Chen, Opt. Lett. 47, 3287–3290 (2022)
Acknowledgements
Part of this work benefits of the PNRR grants SEE-LIFE “StrEngthEning the ItaLIan InFrastructure of Euro-bioimaging” (B53C22001810006, IR0000023), Project “National Center for Gene Therapy and Drugsbased on RNA Technology”, code CN00000041 funded by the European Union—Next Generation EU PNRR MUR—M4C2—Investimento 1.4—Avviso “Potenziamento strutture di ricerca e creazione” di “campioni nazionali di R&S” CUP J33C22001130001, Project NQSTI “NATIONAL QUANTUM SCIENCE AND TECHNOLOGY INSTITUTE”, code PE00000023 funded by the European Union—Next Generation EU PNRR MUR—M4C2—Investimento 1.3—Avviso Creazione di “Partenariati estesi alle università, ai centri di ricerca, alle aziende per il finanziamento di progetti di ricerca di base” CUP J53C22003200005 and the Caribbean grant EU-LAC-2022-156 (4th EU-LAC Joint Call in STI 2022) for AD. The authors acknowledge the support of the ALM (Advanced Light Microscopy) Italian node of EUROBIOMAGING initiative. The authors thank the Nanoscopy group at the Istituto Italiano di Tecnologia and the Biophsyics Group at DIFILAB, Department of Physics, University of Genoa and at IBF (Istituto di Biofisica), CNR, Genova. All the authors thank Michele Oneto for the technical support at NIC@IIT.
Funding
Open access funding provided by Istituto Italiano di Tecnologia within the CRUI-CARE Agreement. Part of this work benefitted from the PNRR grants SEE-LIFE “StrEngthEning the ItaLIan InFrastructure of Euro-bioimaging” (B53C22001810006, IR0000023), Project “National Center for Gene Therapy and Drugsbased on RNA Technology”, code CN00000041 funded by the European Union—Next Generation EU PNRR MUR—M4C2—Investimento 1.4—Avviso “Potenziamento strutture di ricerca e creazione” di “campioni nazionali di R&S” CUP J33C22001130001, Project NQSTI “NATIONAL QUANTUM SCIENCE AND TECHNOLOGY INSTITUTE”, code PE00000023 funded by the European Union—Next Generation EU PNRR MUR—M4C2—Investimento 1.3—Avviso Creazione di “Partenariati estesi alle università, ai centri di ricerca, alle aziende per il finanziamento di progetti di ricerca di base” CUP J53C22003200005 and the Caribbean grant EU-LAC-2022-156 (4th EU-LAC Joint Call in STI 2022) for AD.
Author information
Authors and Affiliations
Contributions
Conceptualization and organization of the review: AD. All authors contributed to the review regarding the different sections, namely: Introduction [AD, PB, CJRS], Mueller matrix [AD, MS, RM, FC, AM, CJRS], The instrument [AD, ALG, PB, FC], Applications in Biophysics and Biomedicine [AD, ALG, AM, FC, PB], A microscope in the machine [AD, LC, PB], Conclusions [AD, CJRS]. The first draft of the manuscript was written by AD and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Diaspro, A., Bianchini, P., Callegari, F. et al. Emerging Mueller matrix microscopy applications in biophysics and biomedicine. Riv. Nuovo Cim. 46, 473–519 (2023). https://doi.org/10.1007/s40766-023-00046-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40766-023-00046-5