Abstract
The existing image semantic segmentation models have low accuracy in detecting tiny targets or multi-targets at overlapping regions. This work proposes a hybrid vision transformer with unified-perceptual-parsing network (ViT-UperNet) for medical image segmentation. A self-attention mechanism is embedded in a vision transformer to extract multi-level features. The image features are extracted hierarchically from low to high dimensions using 4 groups of Transformer blocks with different numbers. Then, it uses a unified-perceptual-parsing network based on a feature pyramid network (FPN) and a pyramid pooling module (PPM) for the fusion of multi-scale contextual features and semantic segmentation. FPN can naturally use hierarchical features, and generate strong semantic information on all scales. PPM can better use the global prior knowledge to understand complex scenes, and extract features with global context information to improve segmentation results. In the training process, a scalable self-supervised learner named masked autoencoder is used for pre-training, which strengthens the visual representation ability and improves the efficiency of the feature learning. Experiments are conducted on cardiac magnetic resonance image segmentation where the left and right atrium and ventricle are selected for segmentation. The pixels accuracy is 93.85%, the Dice coefficient is 92.61% and Hausdorff distance is 11.16, which are improved compared with the other methods. The results show the superiority of Vit-UperNet in medical images segmentation, especially for the low-recognition and serious-occlusion targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Medical images can directly reflect the 2D and 3D morphological characteristics of specific organs and tissues in the human body, with complex structures and diverse contents. Due to the influence of noise, field drift effect, offset deformation, gray value distortion, local body position effect, and tissue movement during image generation, medical images are often blurred. The differences of anatomical structures among individuals also increase the difficulty of feature differentiation. Besides, the boundary of human organs and tissues is fuzzy, and is accompanied by regular or irregular periodic dynamic changes [1, 2]. In the process of medical image segmentation, due to the aforementioned inherent fuzziness of the image, which brings many difficulties to image segmentation [3]. Therefore, it is a challenging task to study efficient and accurate segmentation methods for complex medical image [4, 5].
With the development of deep learning, convolutional neural networks (CNNs) play a dominant role in medical image segmentation. Among various CNN variants, U-Net [6] based on an encoder–decoder architecture shows outstanding performance. The encoder extracts features through continuous downsampling, and then, the decoder uses the output of the encoder for upsampling through skip connection, to obtain features with multiple granularities. Based on U-Net, many models are specially designed for medical image segmentation, such as UNet++ [7], Res-UNet [8], DensueNet [9], R2U-Net [10], KiU-Net [11], and UNet 3++ [12]. Although CNN-based approaches have achieved great success in medical image processing, due to their inherent perceptual bias, each convolution kernel only focuses on a single sub-region of the image, which makes them lose the association of global context information and unable to establish long-term dependency. Especially, in segmentation of medical images such as cardiac magnetic resonance images (MRI), the segmentation accuracy of different organs and tissues is often different. For relatively small tissues such as myocardial or organs with serious occlusion such as the right ventricle, the segmentation accuracy is relatively low, and it is easy to cause excessive segmentation. The reason is that CNN-based methods extract spatial features through continuous pooling operation and convolution, which leads to the reduction of feature resolution and hinders the segmentation of small-target or overlapping-target boundary.
Transformer is a sequence-to-sequence prediction framework. Because of its powerful long-range sequence modeling ability, it has excellent performance in the field of machine translation and natural language processing [13]. The self-attention mechanism in Transformer can dynamically adjust the receptive field according to the input content, effectively establish a global connection among sequential tags, and is superior to convolution operations in modeling long-term dependency. Recently, Transformer has been regarded as an alternative architecture of CNN and has achieved competitive performance in many computer vision tasks, such as image recognition [14, 15], semantic/instance segmentation [16, 17], object detection [18, 19], and image generation [20]. In particular, Detection Transformer adopts a Transformer-based design to construct the first complete end-to-end target detection model. Vision Transformer (ViT) is the first proposed image recognition model based on Transformer, and has achieved the good performance. However, the Transformer-based model has also attracted some attention in medical image segmentation. The TransUNet [21] uses CNN to extract features, and then inputs them into Transformer for long-range context-related modeling. The TransFuse [22] based on ViT attempts to integrate features extracted by Transformer and CNN. To reduce computational cost and combine multi-scale/multi-level features with Transformer, Liu et al. propose a hierarchical Swin Transformer [23] based on a shifted window multi-head self-attention mechanism, which surpasses other advanced methods in image classification and intensive prediction tasks (such as target detection and semantic segmentation). Multi-scale feature representation can bring more powerful performance to visual Transformer. The success of these models shows the great potential of Transformer in medical image segmentation.
In this paper, ViT is combined with an unified-perceptual-parsing network(UPerNet) based on a feature pyramid network and a pyramid pooling module for medical image segmentation. Besides, a self-supervised learner, masked autoencoder, is proposed to pre-train the model, strengthen the visual representation learning ability, and improve the learning efficiency and segmentation accuracy. The main contributions are as follows:
-
(1)
ViT is used to conduct long-range dependency modeling and feature extraction capabilities and a unified-perceptual-parsing network with the multi-level feature fusion capabilities improves the segmentation accuracy for small targets.
-
(2)
A self-supervised learner called masked autoencoder is used for model pre-training, which improves the feature learning efficiency and segmentation accuracy.
The rest of the paper is organized as follows. In section two, an ViT-UperNet is established. The learning algorithm is proposed in section three. In section four, the experiments on medical MRI images are conducted and results are analyzed. Finally, section five concludes this paper and discusses the future work.
ViT-UperNet
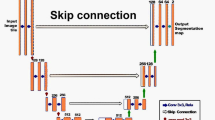
In this section, a self-attention-based vision transformer unified perceptual parsing network (ViT-UperNet) is proposed. It takes ViT as the basic backbone network and UPerNet as the image semantic segmentation module. The input image is divided into small patches, and then, the linear embedding sequence of these patches is input into the network. The ViT network extracts hierarchically features from low dimension to high dimension of the image. After scale transformation, features of different dimensions are used as the input of the UPerNet. A feature pyramid network (FPN) and a pyramid pooling module (PPM) realize the fusion of multi-dimensional features and the association of context information. Then, the final output enters a pixel-level Softmax classifier and a category probability for each pixel is independently generated. The overall structure of ViT-UPerNet is shown in Fig. 1.
In Fig. 1, the left side is the ViT, which is composed of four stages. Each stage contains a different number of Transformer blocks composed of multi-head self-attention (MSA) and MLP layers. The output of each stage is connected to the different scale layer of the FPN module in the right UPerNet after the scale transformation with upsampling or downsampling. The PPM module is inserted after last stage of the ViT and before the first layer of the FPN module. It can ensure that the receptive field of the deep network is not lost, and ensure effective representation of global context prior knowledge. After the multi-scale hierarchical features from the FPN are further fused, a fusion feature map of the same size as the input image is generated by size reduction of upsampling and convolution operations, and the final semantic segmentation result is obtained via a Softmax classifier.
The ViT network based on self-attention
ViT network based on self-attention is used for the hierarchical extraction of image features. It is divided into four stages. Each stage contains a different number of Transformer blocks composed of MSA and MLP layers. The image features are extracted hierarchically from low dimensions to high dimensions. Its main structure is shown in Fig. 2.
In Fig. 2, the input image is first divided into small patches with a certain scale (for example, \(4\times 4\)). Then these patches are expanded in space. It uses trainable linear mapping to transform them to fixed dimensions and achieve the embedding of image patches. Meanwhile, position embedding vectors are added to the patch embedding sequence to retain the position information of 2D image after blocking. The embedded image patches are the input of following four feature-extraction stages. The numbers of Transformer blocks in the four stages are 4, 6, 8, and 6. Notice that Transformer blocks do not change the scale of features.
While extracting features, the output of the last Transformer block in each stage is fused with features of different scales in the FPN after upsampling or downsampling. In particular, the output features of the last stage are used as input to the first layer in the FPN after the global context information fusion of PPM.
In the process of connection with FPN, to fuse the feature maps at same scale extracted by ViT with the feature maps at different scales from FPN, the output feature maps at each stage of ViT are upsampled or downsampled accordingly. For the output features in the first stage and the second stage, the transposed convolution to the corresponding scale is used for upsampling, the output in the third stage remains scale-invariant, and the output in the last stage is downsampled using the maximum pooling operation. Finally, multi-level feature maps corresponding to different layers in FPN are generated (1/4, 11/8, 1/16, and 1/32, respectively) as the input of the previous layer of the horizontal connection of the FPN.
The feature pyramid network (FPN)
FPN is a pyramid form that can naturally use hierarchical features, and generate strong semantic information on all scales. The structure of FPN includes the top–down hierarchical structure and horizontal connection. It integrates the shallow features with high resolution and the deep features with rich semantic information. The structure of FPN is shown in Fig. 3.
In the hierarchical feature-extraction process, high-level features contain more semantic information, while low-level features contain more spatial location information. The FPN upsamples the high-level feature map with stronger semantics, and then horizontally connects the feature to the previous level. FPN can fuse the top-level features with low-level features through upsampling and realizes the rapid construction of feature pyramids with strong semantic information at all scales.
The pyramid pooling module (PPM)
The pyramid pooling module is composed of a set of pooling blocks with different scales, which can better use the global prior knowledge to understand complex scenes and extract features with global context information to improve image recognition or segmentation results, and is an effective global context prior model. The structure of PPM is shown in Fig. 4.
In Fig. 4, PPM has four different pyramid scales. First, the input feature map is pooled to different sizes, and the sizes of layers are \(1\times 1\), \(2\times 2\), \(3\times 3\), and \(6\times 6\). This multi-scale pooling can retain global context information at different scales. Then, to maintain the weight of the global features, \(1\times 1\) convolutions are performed on the pooled results, and the number of channels is reduced to 1/4. Then, the low-dimensional feature map is directly upsampled by bilinear interpolation to obtain the feature map with the same size as the original feature map, and then, the original feature map and the feature map obtained by upsampling are spliced according to channel dimensions. The number of channels obtained is twice that of the original feature map. Finally, \(1\times 1\) convolutions are used to reduce the number of channels to the original number. A feature map with the same size and number of channels as the original feature map is obtained as the output of the pyramid pooling module.
As a hierarchical global structure, PPM further reduces the loss of context information between different scales and regions, and can construct global prior information in the final layer of network.
The information processing flow of the ViT-UPerNet network
The ViT-UPerNet uses the ViT network based on self-attention as the backbone to perform hierarchical extraction and representation of features, and then uses the FPN and PPM modules in UPerNet to fuse the extracted multi-scale features and associate the global context information. Finally, the pixel-level segmentation of the image is achieved through a Softmax classifier. The specific information processing flow is shown in Table 1.
(1) Image input
The three-channel image with the size of \(H\times W\) is input into the ViT-UPerNet. First, the image patches are segmented through the patch segmentation layer, and then, the dimension becomes \((H/16)\times (W/16) \times C\) through the linear embedding layer.
(2) ViT feature extraction
The feature map with the size of \((H/16)\times (W/16) \times C\) goes through four feature-extraction stages. Each stage contains a different number of Transformer blocks, which are 4, 6, 8, and 6, respectively. The processing of the Transformer block does not change the size of the feature map, the channel dimension of the feature map in each phase is 1024, and the number of multi-head in the Transformer block is set to 16. After four stages of feature extraction, a high-level feature map with the size of \((H/4)\times (W/4) \times 1024\) is finally obtained. Meanwhile, the output feature map of each stage is connected to the corresponding FPN layer, and the PPM is used between the top-level feature map and the top-level of the FPN to fuse the global context information.
The outputs of the four stages are, respectively, upsampled or downsampled to maintain consistency with the feature scales of the corresponding levels in the FPN. For the output feature of the first stage (scale is 1/16), first, a transpose convolution with step size of 2 and size of \(2\times 2\) is performed. Then, the layer normalization operation and GELU function activation are performed. Finally, a transpose convolution with step size of 2 and size of \(2\times 2\) is performed, and the scale of the feature map becomes the same as that of the FPN at the same level (1/4). For the output of the second stage, only one transpose convolution with a step size of 2 and a size of \(2\times 2\) is performed to change the scale to 1/8 required by FPN. The characteristic scale of the third stage is the same as that of the corresponding FPN layer, which is directly connected horizontally. The output of the fourth stage uses the maximum pooling of \(2\times 2\) to conduct downsampling, and the characteristic map of scale 1/32 is obtained as the input of the PPM module.
(3) The pyramid pooling module (PPM)
The output from the last layer of ViT uses the maximum pooling for \(2\times \) downsampling to obtain a high-level feature map with the size of \((H/32)\times (W/32)\) and the number of channels of 1024 as the input of the PPM module. First, the average pooling is performed to obtain different scales, and the dimensions of layers are, respectively, \(1\times 1\), \(2\times 2\), \(3\times 3\), and \(6\times 6\). Then, a \(1\times 1\) convolution is performed, and the number of channels is reduced to 1/4. Then, the low-dimensional feature map is upsampled by bilinear interpolation to obtain the feature map with the same size as the original input. The original feature map and the feature map obtained by upsampling are spliced according to channel dimensions. Finally, a \(3\times 3\) convolution is used to reduce the number of channel to 256 as the top-level feature map of the FPN.
(4) The feature pyramid network (FPN)
The size of the PPM is \((H/32)\times (W/32)\times 256\), and the output containing the global scene prior information is used as the top-level feature map of the FPN. From the top to the bottom, each layer of the FPN feature map is first upsampled by double bilinear interpolation. Then, after the feature maps from the same level of ViT are upsampled and downsampled for uniform scale, the channel dimension is converted to 256 by \(1\times 1\) convolution operation. Then, the elements are added with it. Finally, after \(3\times 3\) convolution operations, the fused feature map is obtained by adding the elements as the next layer feature map of FPN. Finally, a multi-level feature map with 256 channels and [1/4, 1/8, 1/16, 1/32] scales is obtained.
(5) Feature fusion
The feature maps from different scales of FPN are upsampled to a unified 1/4 scale using bilinear interpolation. They are spliced on the channel dimension, and reduced to 256 using a \(3\times 3\) convolution operation. Finally, bilinear interpolation is used to upsample the feature map to the original size (\(H\times W\)) and input it to the output layer.
(6) The output
In the output layer, the feature map with the size of \(H\times W\) and the number of channels of 256 is output as a four-dimensional vector of \(H\times W\) through a Softmax classifier. Each dimension corresponds to a class.
The training algorithm
The masked autoencoder pre-training algorithm based on self-supervised learning
Masked autoencoder (MAE) is a scalable self-supervised learner [24]. It randomly masks the patches of the input image and reconstructs the missing pixels. It has an asymmetric encoder–decoder architecture, where the encoder runs only on a subset of visible tiles (without mask tags), and a lightweight decoder for reconstructing the original image from potential representations and mask tags. In the case of high proportion masking, it can efficiently train large models, speed up training, and improve accuracy. The scalable MAE is suitable for high-capacity models with good versatility. Its migration performance in downstream tasks is better than supervised pre-training, and it shows good robustness and adaptability. The structure of MAE is shown in Fig. 5.
MAE is a simple autoencoding method, which reconstructs the original signal according to partial observations of the original signal. Like all autoencoders, it has an encoder that maps the observed signal to the potential representation, and a decoder that reconstructs the original signal from the potential representation. Different from the classic autoencoder, an asymmetric design is adopted, which allows the encoder to operate only on part of the observed signals (unmasked), and a lightweight decoder is used to reconstruct the complete signal from the potential representation and mask marks. The specific process of MAE operation is as follows:
-
(1)
Mask: According to the input format of ViT, the image is divided into regular non-overlapping image patches. Then, the patches of a subset are sampled randomly, and the remaining patches are masked (i.e., removed). The random sampling strategy is to sample random image patches according to uniform distribution without replacement.
-
(2)
MAE encoder: The encoder is the backbone network ViT, but the encoder is only used for visible and unmasked image patches. Same as the standard ViT, the encoder embeds image patches through linear projection, adds position embedding, and then encodes image features from low level to high level through a series of Transformer blocks. Because the encoder only runs on a small part of the whole patches (for example, \(25\%\)), and does not use any masked image patches and mask tags, the computational resources required in the encoding process are greatly reduced.
-
(3)
MAE decoder: The input of MAE decoder is composed of (i) coded output of visible image patches and (ii) mask tags. Each mask tag is a shared learning vector representing the missing patches to be predicted. Meanwhile, the position encoding of each tag is also added to locate the position of the patches. The MAE decoder is only used to perform image reconstruction tasks during pre-training (the encoder generates an image representation for recognition). Therefore, the decoder architecture can be flexibly designed independently of the encoder structure. Then, all tags are only processed by the lightweight decoder, which greatly reduces the pre-training time.
-
(4)
Reconstruction target: MAE reconstructs the input image by predicting the pixel value of each masked patches. Each output element in the decoder represents a pixel value vector of an image patches. The last layer of the decoder is a linear mapping, and the number of output channels is equal to the number of pixel values in an image patch. Finally, the output of the decoder is reshaped to form a reconstructed image.
The loss function of MAE needs to calculate the mean square error between the reconstructed image and the original image in the pixel space, where only the losses of the masked image patches need to be calculated.
The ViT-UPerNet training algorithm
(1) MAE pre-training
The pre-training process of MAE is a self-supervised learning, which takes the image itself as the label, and does not need other segmentation marks. The backbone ViT is used as the MAE encoder, followed by a lightweight decoder. The decoder is composed of 8 consecutive Transformer blocks, and the channel dimension is 512. The last layer of the decoder is a linear mapping, and the number of output channels is equal to the number of pixel values in an image patch, that is \(16\times 16=256\). Finally, the output of the decoder is reshaped to reconstructed image. The loss function of MAE uses mean square error, but only calculates the loss of masked image patches
where \(y_i\) is the real value of a masked pixel, \(y'_i\) is the predicted value of the corresponding pixel, and n is the number of pixels.
(2) The overall adjustment of ViT-UPerNet model
The ViT backbone network pre-trained by MAE is used as the feature extraction module, which is coupled with UPerNet, with the original image as the input, the actual image is divided into truth tags, and supervised learning is used for overall fine-tuning.
In the training stage, the proposed ViT-UPerNet model is trained end-to-end using the objective function. The objective function is calculated by Sorensen–Dice loss and binary cross-entropy function, and a Softmax function is used on the final feature mapping to achieve pixel classification. The calculation formula is
where t is the total number of pixels in each image, y represents the basic true value of the ith pixel, and \(p_i\) represents the confidence score of the ith pixel in the prediction result. In the experiment, \(\alpha =\beta =0.5\), and \(\varepsilon =10^{-6}\).
Experiment
The dataset
The dataset uses cardiac MRI image data from the 2017 Automated Cardiac Diagnosis Challenge (ACDC2017) [25]. All data were collected from clinical examination data of Dijon University Hospital in France, including cardiac MRI image data of 150 patients in the whole cardiac cycle. Each patient also contains some other physiological information, such as height, weight, etc. All cardiac MRI images were collected by two Siemens nuclear magnetic scanners with different magnetic intensities (1.5 T and 3.0 T). After the long-axis sequence of free precession was obtained under retrospective or prospective equilibrium conditions using conventional Steady-State Free Precession (SSFP) method, the short-axis sequence slices covering the left-ventricular region from bottom to top were obtained. The thickness of slices is 5–8 mm, the spacing between slices is 5 mm, and the size of matrix is \(256\times 256\), the field of view is \(300\times 330\,\hbox {mm}^2\), and the spatial resolution is 1.34–1.68 \(\hbox {mm}^2\)/pixel. According to the patient’s condition, 28–40 time-phases were collected to completely cover a complete cardiac cycle.
The ACDC dataset includes not only the MRI images of the complete cardiac cycle, but also part of the individual end-diastolic and end systolic MRI images, as well as the expert manual segmentation and labeling information of the corresponding left ventricle, right ventricle, and myocardial layer.
Because the standard segmentation of the test set of 50 patients included in the ACDC data is not disclosed, and only the end diastolic and end systolic have standard segmentation labels. In this experiment, the end-diastolic and end-systolic sections in the training set containing 100 patients’ data were selected as the experimental dataset for model training and testing. The experimental data set was randomly divided according to the proportion, and consisted of a training set of 70 patients, a verification set of 10 patients, and a test set of 20 patients.
Experimental parameter setting
The experiment is implemented by Python 3.7. The environment is built on the Linux system with CPU Intel Xeon 4214R * 2, and GPU NVIDIA GeForce RTX 2080Ti (12GB) * 3.
In the unsupervised pre-training of MAE, the masking rate of the input image is 50\(\%\), and the random masking method is used. In the training process, the entered \(batch\_Size=12\), and training \(\hbox {epoch}=50\). We select the AdamW optimizer with momentum term [26] to optimize the back propagation of model training with momentum term \(b_1,b_2=0.9,0.95\). The weight decay rate is set to 0.05, the benchmark learning rate is 1.5e−4, and the linear scaling strategy [27] is adopted, and \(lr=base\_ lr/batch\_ size\). We use the cosine decay strategy [28] to perform iterative attenuation of the learning rate.
In the overall training fine-tuning, the entered \(batch\_ Size=12\), and training \(\hbox {epoch}=100\). Similarly, the AdamW optimizer conducts model training optimization, momentum term \(b_1,b_2=0.9,0.99\). The weight decay rate is set to 0.05, the benchmark learning rate is 1e−3, and the hierarchical decay strategy [29] is adopted, the decay rate is 0.75, and the cosine iterative decay strategy is also used for iterative decay of the learning rate.
To measure the segmentation performance of ViT-UPerNet, several commonly used medical image segmentation evaluation indicators are selected, including mean pixel accuracy (MPA), Dice similarity coefficient (Dice), and Hausdorff distance (HD), to evaluate the quality of image segmentation.
Experimental results and discussion
(1) Experimental results
The MAE pre-training algorithm is used to conduct self-supervised pre-training for the ViT backbone network, and 50\(\%\) masking rate is selected to process the input image and output a complete reconstructed image. The reconstruction results of some images are shown in Fig. 6.
It can be seen from Fig. 6 that under the condition of 50\(\%\) masking rate, after MAE pre-training, the ViT backbone can still reconstruct the masked area well, especially for the reconstruction of heart tissue and organ area, which proves that the pre-trained backbone has a strong feature-extraction ability for cardiac MRI images.
We connect the pre-trained ViT backbone network with the UPerNet semantic segmentation framework, and use the training set in the section “The Dataset” for the overall training fine-tune. Then, we verify and analyze the model on the test set. The partial segmentation results of the ViT-UPerNet are shown in Fig. 7. Purple area is the background, yellow area is the left ventricle, green area is the myocardium, and blue area is the right ventricle.
Figure 7 shows the partial segmentation results of the ViT-UPerNet. The first line is the original MRI image, the second line is the truth segmentation, and the third line is the prediction segmentation. It can be seen that the segmentation result predicted is consistent with the segmentation truth value.
(2) Comparative discussion
To prove the effectiveness of the proposed ViT-UPerNet, the image segmentation models are selected for comparative experiments, including ResNet50+DeepLabV3+ [30], ResNet50+UPernet [31], and TransUNet. The encoder of DeepLabv3+ model is a deep convolutional network. The common classification network ResNet50 is used, and then, the spatial pyramid pooling module with dilated convolution is used to introduce multi-scale information. Finally, the decoder module is introduced to further integrate the low-level features and high-level features to improve the accuracy of segmentation boundary. UPerNet is based on the feature pyramid network structure, and uses the pyramid pooling module in PSPNet for the last layer of the backbone network. The model uses features of multiple semantic levels, from scenes, objects, parts, materials to textures, to try to analyze the multi-level visual concept of images at one time. TransUNet adopts a hybrid architecture of CNN and Transformer. It uses CNN to extract fine grained and high-resolution spatial information, and then uses Transformer to associate the global context. All comparison models are end-to-end trained on the same dataset, the results are based on the source code published by the original author, and the comparison algorithm uses default parameters. Three performance indicators, PA, Dice coefficient, and HD, were used to conduct quantitative evaluation on each comparison model. The results are shown in Table 2.
The visualization results of organ segmentation of cardiac MRI images by different contrast models are shown in Fig. 8. From left to right: original MRI image, truth segmentation result, ViT-UPerNet segmentation result, TransUNet segmentation result, ResNet50+UPerNet segmentation result, and ResNet50+DeepLabV3+segmentation result.
It can be seen from Table 2 that the segmentation model proposed in this paper obtained the best measurement index among all comparison algorithms. The final results are 93.85%, 92.61%, and 11.16, which verify the effectiveness. The reason can be attributed to the MAE pre-training mechanism of theViT-UPerNet, the ability of multi-scale feature extraction based on self-attention, and the ability of multi-level feature fusion analysis based on the FPN and PPM.
Figure 8 shows that the segmentation results of the ViT-UPerNet are consistent with the segmentation truth value, and can accurately capture all segmentation targets. However, the ResNet50+UPerNet and ResNet50+DeepLabV3+methods based on pure convolution, to some extent, are over segmented, especially for the right ventricle with large morphological changes and the myocardial layer with low pixel occupancy. In contrast to the TransUNet based on Transformer network, the segmentation result of ViT-UPerNet is more smooth at the boundary, especially for the left ventricle and left-ventricular myocardial layer with overlapping positions, showing good robustness.
(3) The ablation experiment and discussion
To further verify the role of MAE pre-training in improving the segmentation accuracy, a comparative experiment was designed with and without MAE pre-training. The comparison results are shown in Table 3.
It can be seen from Table 2 that the ViT-UPerNet with MAE pre-training exceeds the model without pre-training in every index, which fully proves that the MAE pre-training method plays a positive role in improving the feature-extraction ability and pixel segmentation accuracy. Figure 9 shows the Dice score curve of MAE ablation experiment training process.
It can be seen from Fig. 9 that the segmentation performance of the MAE pre-trained model can quickly reach a higher level during the overall parameter fine-tuning training process, and the segmentation performance fluctuates less and is more stable. It proves that the MAE pre-training method can improve the training efficiency and segmentation accuracy, and enhance the stability and robustness of the model.
Conclusion
This paper proposes a self-attention-based Vision Transformer with unified-perceptual-parsing network, dubbed as ViT-UperNet, for solving the problem of difficult segmentation of small and overlapping targets in medical images. The Vision Transformer network based on the self-attention is used as backbone for multi-level feature extraction, and the unified perceptual parsing network based on an FPN and a PPM is used for multi-scale context information fusion. We combine the feature-extraction capability of the Vision Transformer with the multi-level feature fusion capability of the unified perceptual parsing network to enhance the boundary division ability to small or overlapping targets. Comparative experiments verify the effectiveness of ViT-UperNet for small-sized/overlapping tissue and organ segmentation. It has shown the potential to effectively learn the critical anatomical relationships represented in medical images.
References
Suganyadevi S, Seethalakshmi V, Balasamy K (2022) A review on deep learning in medical image analysis. Int J Multimed Inf Retr 11(1):19–38
Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi AK (2022) Medical image segmentation using deep learning: a survey. IET Image Proc 16(5):1243–1267
Alagarsamy S, Govindaraj V et al (2023) Automated brain tumor segmentation for MR brain images using artificial bee colony combined with interval type-II fuzzy technique. IEEE Trans Ind Inf 19(11):11150–11159
Xun S, Li D, Zhu H, Chen M, Wang J, Li J, Chen M, Wu B, Zhang H, Chai X et al (2022) Generative adversarial networks in medical image segmentation: a review. Comput Biol Med 140:105063
Lin A, Chen B, Xu J, Zhang Z, Lu G, Zhang D (2022) Ds-transunet: dual swin transformer u-net for medical image segmentation. IEEE Trans Instrum Meas 71:1–15
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M (2023) Swin-unet: Unet-like pure transformer for medical image segmentation. In: Computer Vision—ECCV 2022 Workshops: Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part III. Springer, pp 205–218
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) Unet++: a nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support: 4th international workshop, DLMIA 2018, and 8th international workshop, ML-CDS 2018, held in conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. Springer, pp 3–11
Xiao X, Lian S, Luo Z, Li S (2018) Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th international conference on information technology in medicine and education (ITME). IEEE, pp 327–331
Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A (2018) H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging 37(12):2663–2674
Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK (2018) Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv preprint arXiv:1802.06955
Valanarasu JMJ, Sindagi VA, Hacihaliloglu I, Patel VM (2020) Kiu-net: towards accurate segmentation of biomedical images using over-complete representations. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 363–373
Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y, Han X, Chen Y-W, Wu J (2020) UNet 3+: a full-scale connected UNet for medical image segmentation. In: ICASSP 2020-2020 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1055–1059
Gillioz A, Casas J, Mugellini E, Abou Khaled O (2020) Overview of the transformer-based models for NLP tasks. In: 2020 15th conference on computer science and information systems (FedCSIS). IEEE, pp 179–183
Meng L, Li H, Chen B-C, Lan S, Wu Z, Jiang Y-G, Lim S-N (2022) Adavit: adaptive vision transformers for efficient image recognition. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 12309–12318
Zhang Q, Xu Y, Zhang J, Tao D (2023) Vitaev2: vision transformer advanced by exploring inductive bias for image recognition and beyond. Int J Comput Vis 131:1141–1162
Wang Y, Xu Z, Wang X, Shen C, Cheng B, Shen H, Xia H (2021) End-to-end video instance segmentation with transformers. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 8741–8750
Cheng B, Misra I, Schwing AG, Kirillov A, Girdhar R (2022) Masked-attention mask transformer for universal image segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 1290–1299
Han G, Ma J, Huang S, Chen L, Chang S-F (2022) Few-shot object detection with fully cross-transformer. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 5321–5330
Fan L, Pang Z, Zhang T, Wang Y-X, Zhao H, Wang F, Wang N, Zhang Z (2022) Embracing single stride 3d object detector with sparse transformer. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 8458–8468
Zhang B, Gu S, Zhang B, Bao J, Chen D, Wen F, Wang Y, Guo B (2022) Styleswin: transformer-based gan for high-resolution image generation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 11304–11314
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) Transunet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306
Zhang Y, Liu H, Hu Q (2021) Transfuse: Fusing transformers and cnns for medical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 14–24
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 10012–10022
He K, Chen X, Xie S, Li Y, Dollár P, Girshick R (2022) Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 16000–16009
Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng P-A, Cetin I, Lekadir K, Camara O, Ballester MAG et al (2018) Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging 37(11):2514–2525
Yong H, Huang J, Hua X, Zhang L (2020) Gradient centralization: a new optimization technique for deep neural networks. In: European conference on computer vision. Springer, pp 635–652
He T, Zhang Z, Zhang H, Zhang Z, Xie J, Li M (2019) Bag of tricks for image classification with convolutional neural networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 558–567
Cubuk ED, Zoph B, Mane D, Vasudevan V, Le QV (2019) Autoaugment: learning augmentation strategies from data. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 113–123
Cui Y, Che W, Liu T, Qin B, Yang Z (2021) Pre-training with whole word masking for Chinese Bert. IEEE ACM Trans Audio Speech Lang Process 29:3504–3514
Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European conference on computer vision (ECCV), pp 801–818
Xiao T, Liu Y, Zhou B, Jiang Y, Sun J (2018) Unified perceptual parsing for scene understanding. In: Proceedings of the European conference on computer vision (ECCV), pp 418–434
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruiping, Y., Kun, L., Shaohua, X. et al. ViT-UperNet: a hybrid vision transformer with unified-perceptual-parsing network for medical image segmentation. Complex Intell. Syst. 10, 3819–3831 (2024). https://doi.org/10.1007/s40747-024-01359-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-024-01359-6