Abstract
Recently, graph contrastive learning (GCL) has achieved remarkable performance in graph representation learning. However, existing GCL methods usually follow a dual-channel encoder network (i.e., Siamese networks), which adds to the complexity of the network architecture. Additionally, these methods overly depend on varied data augmentation techniques, corrupting graph information. Furthermore, they are heavily reliant on large quantities of negative nodes for each object node, which requires tremendous memory costs. To address these issues, we propose a novel and simple graph representation learning framework, named SimGRL. Firstly, our proposed network architecture only contains one encoder based on a graph neural network instead of a dual-channel encoder, which simplifies the network architecture. Then we introduce a distributor to generate triplets to obtain the contrastive views between nodes and their neighbors, avoiding the need for data augmentations. Finally, we design a triplet loss based on adjacency information in graphs that utilizes only one negative node for each object node, reducing memory overhead significantly. Extensive experiments demonstrate that SimGRL achieves competitive performance on node classification and graph classification tasks, especially in terms of running time and memory overhead.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Recently, graph representation learning has emerged as an effective way to analyze graph-structured data, which mostly relies on graph neural networks (GNNs). Graph representation learning aims to learn high-quality embeddings that preserve the topological information of graphs and node properties. However, most prior GNN models, such as GCN [20] and GAT [36], still follow the supervised learning paradigm that requires plenty of labeled training data. Obtaining annotated labels for graphs is often prohibitively expensive or impossible, limiting their applicability to real-world scenarios. For example, producing labels for molecular graphs usually requires costly calculations. To address this drawback of supervised learning, self-supervised learning (SSL) has been presented as a method that can effectively utilize vast amounts of unlabeled data during training.
Motivated by the great achievement of SSL based on contrastive learning in computer vision (CV) [4, 5, 13] and natural language processing (NLP) [7, 10], there has been substantial research attention given to the application of SSL to graph domains. Inspired by Deep InfoMax [13], DGI [37] first employed contrastive learning in the graph domain. Recently, major graph contrastive methods have been adapted from the SimCLR [4], which follow a dual-channel network (i.e., the Siamese network) and rely on data augmentations. According to the SimCLR [4], stronger data augmentations used for images can bring more benefits. To this end, Grace [49] and GraphCL [47] proposed several kinds of data augmentations applicable to graphs. Moreover, GCA [50] introduced a kind of adaptive augmentation used for graphs, which was a more flexible data augmentation strategy for graph contrastive learning (GCL).
Although the aforementioned methods have significant performance in graph representation learning, they still suffer from the following limitations: Firstly, existing GCL methods heavily depend on data augmentations. Due to the complexity of graphs, the strong data augmentation techniques available for graphs are generally non-trivial. At the same time, data augmentation usually damages graph information, including node features and structure information. In addition, there is no standard data augmentation technique for graphs that can effectively adapt to every task. Thus, the various kinds of data augmentations are usually selected according to the specific scenarios, restricting their scope of application. Then, after obtaining two augmented views via data augmentations, these pairs of augmented representations have to be sent to the dual-channel network, followed by the projection head, for training. Also, this process increases network complexity. Finally, for most existing graph contrastive methods, each object node has been compared with large quantities of negative nodes that contain every node in the graph except for the node itself, which raises computation and memory costs. Albeit a recent work by BGRL [34] aims to throw away negative nodes during training, it calls for an elaborate asymmetric Siamese network. From these perspectives, we naturally wonder whether one method can overcome these limitations as simply as possible.
In this paper, we propose a simple self-supervised graph representation learning framework, named SimGRL, to solve all of the above limitations. Firstly, unlike existing GCL methods with the two-encoder network, we only employ an encoder based on graph neural networks, aggregating information from node features and topological structure. Then, we propose a distributor capable of generating triplets to obtain contrastive views of nodes (positive pairs), which consist of nodes and their corresponding neighbor nodes. Owing to the distributor only relying on adjacency information, our proposed method can avoid using diverse data augmentations to gain contrastive views of nodes. Furthermore, we utilize a triplet loss based on adjacency information, which only focuses on a negative node for every object node. Compared with the losses of the aforementioned graph contrastive methods, this type of loss requires much less memory overhead. In the meantime, this triplet loss allows our model to maximize the similarity between positive pairs, promoting the formation of high-quality features. We illustrate the main difference between our method and the prior graph contrastive methods in Table 1.
To sum up, our main contributions are summarized as follows:
-
We propose a simple and novel SSL paradigm for graph representation learning, called SimGRL. SimGRL can efficiently work with a single-channel encoder compared with the prior graph contrastive methods that have a dual-channel encoder.
-
We present a distributor that generates triplets as contrastive views of nodes, allowing SimGRL to perform well without data augmentations.
-
We design a triplet loss based on adjacency information that only leverages a negative node for every object node, considerably reducing memory overhead.
-
We empirically show that SimGRL achieves competitive performance on both node classification tasks and graph classification tasks, especially on running time and memory overhead.
The rest of the paper is organized as follows: Sect. 2 discusses the related work. Section 3 introduces the problem definition of self-supervised graph representation learning. Section 4 describes the proposed SimGRL framework. Section 5 presents an extensive experimental analysis of the proposed method. Finally, Sect. 6 concludes the paper.
Related work
Graph representation learning
Graph representation learning has provided a powerful capacity to leverage the rich information of graph-structured data, efficiently applied in various domains [29, 39, 45]. As a result, numerous approaches to learning graph representations have been proposed in recent years.
For example, inspired by word2vec [49], DeepWalk [28] and node2vec [11] utilize the skip-gram model to generate node representations by adopting random walks across nodes. However, this kind of method cannot fully leverage node features [46] and contains a multi-step pipeline that requires individual optimization for each step, such as random walk generation and semi-supervised training [20].Graph kernel approaches [21, 23] aim to decompose the graph into substructures and use appropriate kernel functions for measuring graph similarity. The mapping function that helps graph kernels embed graphs or nodes into vector spaces is deterministic [41]. Even worse, the computation of all kernel values for graph kernels takes quadratic time [26]. Graph auto-encoders [19, 27] seek to train an encoder that maps input representations to intermediate representations and a decoder that tries to recover input representations based on intermediate representations. Their key is to minimize the reconstruction error between original input representations and recovered input representations [41].
Graph neural networks (GNNs) are popular methods for learning graph representations [16]. In the GNNs framework, each GNNs layer (i.e., a learnable mapping function) intends to obtain the embedding of a node by combining data from the node’s neighbors based on the non-linear transformation and the aggregation function [2]. Compared to DeepWalk and Node2Vec, it is a single-step training framework that can effectively use node features. In addition, the primary benefit of GNN over graph kernels is that their complexity scales linearly with the number of samples [26]. Moreover, graph neural networks can be considered the encoder component of graph auto-encoders. Hence, rather than employing the others, we choose the graph neural network as our encoder because it is more effective and efficient.
Self-supervised graph representation learning
Recently, motivated by the significant success of self-supervised learning (SSL) methods in CV and NLP, many researchers have sought to apply SSL methods to graph domains. Following the idea of Deep Infomax [13], DGI [37] and InfoGraph [33] intend to maximize the mutual information (MI) between two different levels of representation. Inspired by the SimCLR [4], a series of SSL methods for graph representation learning have been proposed, achieving state-of-the-art performance in graph representation learning. For example, Grace [49] introduced two specific schemes to obtain two augmented views of graphs, including removing edges and masking features. GraphCL [47] proposed four types of graph augmentation techniques, such as node dropping, edge perturbation, and subgraph. Moreover, GCA [50] provided a kind of adaptive data augmentation used in graphs. Their key idea is to maximize the similarity between two augmented views of graphs sent to the dual-channel network.
However, the above methods still follow the dual-channel network architecture and heavily depend on diverse data augmentations. At the same time, these methods also heavily rely on massive negative nodes. In contrast to them, our proposed method avoids using data augmentations and employs a single-channel network as our framework. Most importantly, we only employ a negative node for every object node, considerably decreasing memory overhead.
Triplet loss
As a popular method in supervised visual representation learning, triplet loss attempts to learn the discriminative feature representations, which is designed in such a way as to pull similar sample pairs from the same class closer and push away dissimilar sample pairs from different classes. Due to the effectiveness of triplet loss, it has been widely applied in various computer vision tasks in a supervised manner, such as image retrieval [24], face recognition [9], person re-identification [48], and object tracking [8].
However, in contrast to the above methods based on supervised learning in computer vision, we design a new triplet loss for self-supervised graph representation learning, which is used as our objective in this work. Due to the original triplet loss that fails to address self-supervised tasks, our loss is based on the adjacency information of graphs for adapting self-supervised tasks, instead of using the label information.
Problem definition
In this section, we formally define the problem of self-supervised graph representation learning. Self-supervised graph representation learning aims to obtain good high-level node representations without leveraging any label information about nodes.
To be specific, suppose we are given a set of features, \(\varvec{X}\)=\(\left\{ \varvec{x}_{1},\varvec{x}_{2},...,\varvec{x}_{N}\right\} \in {R^{N\times {M}}}\), where N is the number of nodes in the graph and \(\varvec{x}_{i}\in {R^{M}}\) is a M dimensional real-value attribute vector associated with node i. In addition, we are provided the graph-structured information of the undirected graph in the form of an adjacency matrix \(\varvec{A}\in {R^{N\times {N}}}\). \(\varvec{A}_{ij}=1\) if node i and node j are connected and \(\varvec{A}_{ij}=0\) otherwise. We intend to learn an encoder \(f(\cdot )\), which maps the original node feature into a high-level representation space based on the information of node and topological information of the graph, such that \(f(\varvec{X},\varvec{A})=\varvec{H}=\left\{ \varvec{h}_{1},\varvec{h}_{2},...,\varvec{h}_{N}\right\} \in {R^{N\times {d}}}\) represents the high-level representation matrix, where \(\varvec{h_{i}}\in {R^{d}}\) denotes the high-level representation for each node i and d is the embedding dimension. Then, these high-level representations can be exploited for downstream tasks, such as node classification and graph classification tasks.
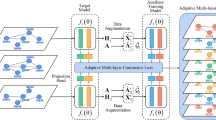
The overall framework of our proposed method, SimGRL. We first use an encoder based on a graph neural network to aggregate information from node features and topological structure. Then, a distributor is used to generate triplets according to the guidance of adjacency information, which contain anchor nodes, positive nodes, and negative nodes. \(SP(\cdot )\) and \(SN(\cdot )\) are used to generate positive nodes and negative nodes respectively. Finally, we train our model with the triplet loss, which can result in the model output having a larger inter-class variation and a smaller intra-class variation
Proposed approach
In this section, we propose a simple graph representation learning framework, called SimGRL. The overall architecture of our proposed method is shown in Fig. 1. In what follows, we will introduce its main components in detail.
Encoder
The core component of the prior graph contrastive methods is the Siamese network with two encoders. By obtaining two augmented views of graphs via data augmentations, these augmented views of graphs (positive pairs) have to be sent to the Siamese network to maximize the agreement between positive pairs. However, we argue that data augmentation is a needless component in our work, and thus there is no need to use the two-encoder network. Hence, we employ only one encoder based on the vanilla graph neural network(GCN) [20] in our proposed network, which is used to gather the information from nodes’ features and topological structure. The encoder f aims to map the node features into high-level space to generate high-quality features. Formally, we formulate the encoder architecture as follows:
where \(\varvec{\hat{A}}=\varvec{A}+\varvec{I}_N\) denotes the adjacency matrix with added self-connections, \(\varvec{I}_N\) is the identity matrix, \(\varvec{\hat{D}}\) represents the degree matrix, \(\varvec{W}\) denotes a trainable weight matrix, \(\varvec{X}\) is the initial feature matrix of nodes, and \(\sigma (\cdot )\) denotes an activation function. In practice, we select the \(PReLU(\cdot )\) as the activation function. \(f(\cdot )\) represents the encoder. \(\varvec{H}\in {R^{N\times {d}}}\) denotes the output node representations through the encoder, where N is the number of nodes and d is the embedding dimension of nodes.
Distributor
Utilizing the rich topological information of graphs used in self-supervised learning is a more natural choice than leveraging data augmentations to corrupt the original information of graphs. Inspired by the triplet network [14], we intend to design a distributor that can generate triplets to form contrastive views of nodes, which replaces data augmentations. However, the original definition of triplets relies on label information, which fails to generate triplets in unsupervised tasks.
To this end, we follow the assumption from the vanilla graph neural network [20]. The assumption is that connected nodes are likely to share the same labels, which is the key to the vast majority of graph neural networks. Additionally, based on the assumption of community detection[17], the neighbors have similar representations. Thus, we extend the idea of triplets to graph domains based on the above assumptions. Firstly, we select the first-order neighbors of anchor nodes (i.e., the nodes themselves) as positive nodes, which compensates for the information provided by the labels. Then the straightforward idea for selecting negative nodes is to select nodes that are not directly connected to anchor nodes.
Practically, as for the positive node, we leverage a positive selector \(SP(\cdot )\) to obtain the positive node, which is based on the guidance from the adjacency matrix. Then,for simplicity, we use a negative selector \(SN(\cdot )\) that randomly and uniformly selects a node from the whole graph as the negative node. The positive selector and the negative selector constitute the distributor together. Using the distributor, we can obtain the contrastive views of nodes that contain the positive pairs constituted from the anchor nodes and corresponding positive nodes and the negative pairs constituted from the anchor nodes and corresponding negative nodes. Finally, the positive selector and the negative selector can be formulated as the following expressions:
where \(Random(\cdot )\) is a function that randomly and uniformly selects an element from a set, \(\varvec{A}_{i\cdot }\) is the i-th row vector of the adjacency matrix \(\varvec{A}\), j is the sequence number of first-hop neighbors belonged to node \(v_{i}\), \(\mathcal V\) is a set of all the nodes in a graph, and \(v_{k}\) denotes the k-th node of \(\mathcal V\).
Triplet loss
The prior graph contrastive methods usually utilize a contrastive objective that aims to maximize the similarity between the two augmented views of the same node. However, this type of objective heavily relies on massive negative nodes that contain every node except for the object node, requiring huge memory costs. At the same time, the composition of negative nodes in their works may be unreasonable. Specifically, they treat all nodes as negative nodes except for the object node to easily obtain massive negative nodes, which probably contain a lot of negative nodes that have the same label as the object node. To alleviate this issue, intuitively, we provide a negative sampler that only samples an unconnected node for object nodes as their negative nodes, which reduces the likelihood of sampling nodes with the same labels as object nodes. Moreover, we empirically find that randomly sampling nodes for object nodes is also effective and simple, and this will be discussed in detail in Sect. 5.6.
Then, we introduce a triplet loss based on adjacency information (i.e., the distributor) as our objective. Compared with the contrastive objective, this objective utilizes only one negative node for every object node. By achieving this objective, we can also enforce our model to maximize the similarity between the node embeddings of contrastive views and minimize the agreement between the embeddings of negative pairs. Specifically, the triplet objective function of an object node is defined as:
where \(\varvec{h}_{i}^{a}\) is the embedding of the object node, \(\varvec{h}_{i}^{p}\) denotes the embedding of the positive node(i.e., \(\varvec{h}_{j}\)), \(\varvec{h}_{i}^{n}\) represents the embedding of the negative node (i.e., \(\varvec{h}_{k}\)), j and k can be obtained by \(SP(\cdot )\) and \(SN(\cdot )\). M is the margin value, N is the number of nodes in the graph, and \(ReLU(\cdot )\) denotes the activation function. Then, the overall training objective can be formulated as the following loss function:
In a nutshell, in each iteration, SimGRL first utilizes an encoder to aggregate information about neighbors. Then, it employs a distributor to generate graph triplets. Finally, the parameters of the learning matrix \(\varvec{W}\) are updated by optimizing the triplet loss. The overall algorithm is summarized in Algorithm 1.

Experiments and analysis
In this section, we conduct extensive experiments to evaluate the proposed method on eight public datasets guided by four research questions.
-
Q1: How does SimGRL compare to state-of-the-art methods on the node classification and graph classification tasks in terms of accuracy?
-
Q2: Is the proposed SimGRL framework efficient in comparison to state-of-the-art methods in terms of running time and memory overhead?
-
Q3: How does SimGRL perform when utilizing different distributor selectors?
-
Q4: Is the proposed SimGRL framework robust when noisy nodes invade triplets?
Datasets
To make a fairly comprehensive comparison, we conducted extensive experiments on various widely used benchmark datasets.
For the node classification task, we use three classical citation networks, Cora, Citeseer, and Pubmed [30], where nodes features are the bag of word representations of documents, edges associated with non-directed citations, and nodes labels stand for the genre of documents. Additionally, we select two large-scale datasets from Open Graph Benchmark [15], i.e., ogbn-arxiv and ogbn-products. ogbn-arxiv denotes a paper citation network of ARXIV papers, and ogbn-products represents an Amazon product co-purchasing network.For the graph classification task, we conducted experiments on the following datasets: Firstly, the MUTAG [22] dataset is formed by chemical compounds that are considered graphs, and their labels correspond to their mutagenic effect on a bacterium. As for the IMDB-binary dataset and the IMDB-multi dataset [43], their nodes represent actors and actresses, and their edges denote that they appear in the same movie. Moreover, the label information of movie networks relies on the genre of movies.
Additionally, we require a metric to evaluate the homophily of each dataset, since our proposed method mainly depends on the homophily assumption that connected nodes are likely to share the same labels. Here, we define an edge homophily ratio as our metric. Formally, the edge homophily ratio is defined as follows:
where E is an edge set, (u, v) denotes a pair of nodes, and \(y_u\) represents the label of the node u. The edge homophily ratio \(h_r\) is the fraction of edges connecting two nodes of the same class. The closer the edge homophily ratio is to one, the stronger the homophily for graphs. It should be noted that this definition is mostly applicable to the datasets used in node classification tasks, and thus we only present the edge homophily ratios of Cora, Pubmed, and Citeseer. The detailed statistics for the datasets are summarized in Tables 2 and 3 respectively.
Experimental settings
For the node classification task, we utilize a linear classifier to evaluate the performance of the learned node representations, which follows the previous state-of-the-art approach DGI [37]. We then report the mean classification accuracy along with the standard deviations after 10 runs of employing the linear classifier.
For the graph classification task, we follow the same evaluation procedure as InfoGraph [33] that assesses the classification performance by leveraging a linear SVM. Moreover, we also use 10-fold cross-validation accuracy with the standard deviation to make a fair comparison.
The experiments on large-scale datasets (i.e., ogbn-arxiv, ogbn-products) were conducted on a machine with one NVIDIA A100 GPU card (40 GB of RAM), and the rest of the experiments were conducted on a machine with one NVIDIA 2080Ti GPU card (11 GB of RAM).All the methods were implemented in Python 3.8 with Pytorch. The hidden layer dimension is 512 as the encoder output dimension. We use the Adam learning algorithm as our optimizer. The learning rate of the optimizer is 0.01 for all datasets. The \(L_{2}\) regularization value is only set 0.0005 on the citeseer dataset. To search for proper margin values, we conduct experiments with different margin values. The results are shown in Fig. 2. Empirically, the margin value M is set as 1 for Cora, 1 for Citeseer, 0.2 for Pubmed, 0.2 for MUTAG, 1 for IMDB-BINARY, 0.6 for IMDB-MULTI, 0.2 for ogbn-arxiv, and 1 for ogbn-products.
Baselines
Node classification. For unsupervised learning methods, we have selected nine methods, including Raw features, DeepWalk [28], GAE [19], DGI [37], GRACE [49], GraphCL [47], GCA [50], BGRL [34] and SelfGNN [18]. These approaches cover most unsupervised learning ideas in the graph field, including random-walk, autoencoder, and contrastive learning. And the rest of the graph kernel methods that are usually used in graph classification are employed in the following comparison of the graph classification task. Aside from unsupervised approaches, we also use five popular semi-supervised methods for comparison, including Planetoid [6], Chebyshev [44], GCN [20], SGC [40], and GAT [36].
Graph classification. For a comprehensive comparison, we select three kinds of methods, including graph kernel approaches, supervised methods, and unsupervised methods. Firstly, we compare our proposed method with five popular graph kernel approaches, including the shortest path kernel (SP) [3], Graphlet kernel (GK) [32], Weisfeiler-Lehman sub-tree kernel (WL) [31], deep graph kernel (DGK) [43] and the multi-scale Laplacian kernel (MLG) [21]. In addition, we also utilize 5 supervised GNNs for comparison: GraphSAGE [12], GCN [20], GAT [36], and two variants of GIN [42]: GIN-0 and GIN-\(\epsilon \). Finally, we compare the results with six unsupervised methods: Random Walk [35], Node2vec [11], Sub2vec [1], Graph2vec [25], InfoGraph [33], and HTC [38].
Comparison with baselines (Q1)
The ultimate purpose of self-supervised representation methods is to learn good high-level representations for downstream tasks, such as node classification and graph classification. Thus, we conduct extensive experiments on node classification and graph classification to evaluate the effectiveness of our proposed method. The results are represented in Tables 4, 5, 6, 7, 8.
First, we aim to analyze the results of node classification tasks on small datasets, i.e., Cora, Citeseer, and Pubmed. As shown in Table 4, we can observe that our proposed method outperforms the supervised baselines across all datasets. For example, SimGRL obtains 1.8% improvement, 0.2% improvement, and 1.7% improvement on Cora, Citeseer, and Pubmed over the classically supervised graph neural network (GAT), respectively. Due to effectively leveraging massive unlabeled samples during training, our proposed self-supervised method can outperform the classical supervised graph neural networks. From Table 5, we can observe that our method has state-of-the-art performance compared to unsupervised methods. It strongly demonstrates that our method can effectively extract the graph structure information as our self-supervised target.
Second, to further evaluate the scalability and performance of our proposed model, we chose two popular large-scale datasets from Open Graph Benchmark [15], which are ogbn-arxiv and ogbn-products. The results are reported in Table 6. First, on the ogbn-arxiv, we can observe that SimGRL outperforms other baselines except for GCN. This is due to the poor homophily ratio of the ogbn-arxiv, which prevents SimGRL from efficiently utilizing the homophily information. Additionally, note that the ogbn-products has a higher homophily ratio, SimGRL consistently performs better than baselines by considerable margins. Furthermore, based on our computing resources, most self-supervised baselines struggle to run on such large-scale datasets, however, SimGRL can. This implies SimGRL’s good scalability.
Third, from Table 7, we find that SimGRL is superior to MLG on the MUTAG with 1.2% improvements, WL on the IMDB-BINARY with 2.2% improvements, and WL on the IMDB-MULTI with 4.4% improvements, respectively. The great performance verifies the superiority of our method compared with graph kernel methods. From Table 8, although SimGRL fails to completely exceed all the supervised methods, it still has competitive improvements in several supervised methods. For instance, compared with GraphSAGE, it improves 4%, 2.2%, and 0.5% on the MUTAG, IMDB-BINARY, and IMDB-MULTI datasets, respectively, which demonstrates that our proposed self-supervised method has the potential to surpass the supervised methods in graph classification tasks. Moreover, from Table 9, SimGRL achieves 1.2% and 0.9% improvements over the state-of-the-art unsupervised graph contrastive method (HTC) on IMDB-BINARY and IMDB-MULTI datasets, respectively. Although SimGRL has competitive results on graph classification tasks, it cannot obtain the best performance on these tasks compared to the most supervised methods and several self-supervised methods. One possible explanation is that the homophily assumption is not universally valid in graph-level tasks (e.g., molecular graphs like MUTAG adopted in the experiments).
All in all, on the node classification and graph classification tasks, it is apparent to note that SimGRL is competitive with not only unsupervised but also supervised methods. These empirical performance improvements demonstrate the superiority of our proposed framework.
Comparison of running time and memory overhead (Q2)
The main goal of our method is of using a simple framework and avoiding massive negative nodes for contrast so that we significantly reduce the computational and memory cost. In Table 1, we qualitatively analyze the advantages of the SimGRL model. The proposed model simplifies the network structure of encoders from two channels to one channel. More importantly, it avoids the consumption of computing resources caused by massive negative nodes. For instance, given a graph contains N nodes, the prior methods require \(N-1\) negative nodes to compute for every object node, while we only need one negative node to calculate for every object node. Furthermore, they need \(O(N^{2})\) memory to store the negative samples, while we only require O(N) memory. Therefore, we used the same software and hardware configuration environment to verify our performance in terms of running time and memory overhead. We compare our method with several state-of-the-art graph contrastive methods, including DGI [33], GraphCL [47], SelfGNN [18], and GCA [50]. Table 10shows the result of the comparison of both running time and memory overhead.
As shown in Table 10, we can observe that our method achieves the minimum of both running time and memory overhead. For instance, on the Cora dataset, the prior best method (DGI) needs 10 s in terms of the running time, while our method only requires 0.7 s. As for the memory overhead, the prior best method (GCA) uses 1200 MB of memory on the Cora dataset, while our method only needs 700 MB. Hence, it convincingly proves that our proposed method can reduce memory overhead and running time considerably compared with the state-of-the-art graph contrastive methods.
Furthermore, more negative examples will be helpful for contrastive methods in computer vision, according to the SimCLR. Thus, we further explore the impact of the number of negative nodes on accuracy and memory overhead. We fixed the number of positive nodes, and the range of negative nodes’ numbers is determined by computing resources. The experimental results are reported in Fig. 3. We can obtain the following observations: First, the memory overhead significantly increases when the number of negative nodes grows. Second, the performance of the model will be improved with more negative nodes. Third, this improvement is limited. As can be seen, after more than 200 negative nodes, the accuracy curves for Cora and Citeseer start to converge. Additionally, when the number of negative nodes is greater than 200, the Pubmed accuracy curve still tends to rise. The potential reason is that the larger datasets have more nodes, and thus they may need more negative nodes to obtain discriminative features that will improve the model’s performance. Finally, since increasing the number of negative nodes will significantly increase the memory overhead, we can conclude that the cost of the improvement is significant.
Effect of different selectors(Q3)
We further analyze the effects of different positive selectors and negative selectors. For a fair comparison, the experiment setup follows the same aforementioned settings except for the selector. For this analysis, we devise several different strategies for the positive selector (SP) and the negative selector (SN). Firstly, we employ a mean SP that obtains the average representation of all the first-order neighbors as the positive node. Secondly, we utilize a random SP that randomly selects a positive node from the first-order neighbors. Thirdly, we devise an unconnected SN that only selects a node unconnected with the object node as the negative node. Finally, we use a random SN that randomly and uniformly selects a node from the whole graph as the negative node.
As we can see in Fig. 4, the random SP performs better than the mean SP, but there is only a slight gap between them. The results indicate that the proposed method is not too sensitive to the different selection strategies of the first-order neighbors. From Fig. 5, we note that the performance of the unconnected SN is also close to the performance of the random SN. One possible reason is that the number of unconnected nodes for an object node is far larger than the number of its neighbors, and thus the results obtained by the two SN appear to be very close.
Impact of the intrusion of noise (Q4)
For analyzing the robustness of our method, we design an experiment that seeks to mix nodes with the opposite type of nodes, simulating the intrusion of noise. The results are presented in Fig. 6. We define a mix rate, which is the proportion of the replaced nodes in the total nodes. Two mixed situations are considered. Firstly, we define P2N, which represents the positive mix type, which replaces positive nodes with negative nodes. Secondly, we define N2P, which denotes the negative mix type, which replaces negative nodes with positive nodes.
We may make the following observations by analyzing the figure: First, as it can be seen from Fig. 6, the accuracy gradually declines as the mix rate increases. We analyze the reason for this by noting that the learning ability of the model tends to degenerate when the difference between positive and negative nodes gradually weakens. Generally, its performance has only a little fluctuation when the mix rate is below thirty percent. This finding suggests that our proposed method can still perform well with a certain range of noise. Moreover, on the Cora and Pubmed datasets, we observe that our method can still perform well when the effect of noise is low (before the mixing rate is lower than 50%), proving that it is robust. However, the model’s performance rapidly deteriorates when the mixing rate is greater than 50% because of an imbalance between positive and negative samples. On Citeseer, because the average number of neighbors is low, it is easy to be bothered by noise. As a result, its performance fluctuates more than the other datasets at lower mixing rates, as can be shown. Furthermore, owing to sampling nodes from the whole graph as negative nodes, we can observe that the performance in N2P is better than in P2N.
T-SNE visualization
To demonstrate the superiority of our model, we display the node embeddings of the Cora dataset by SimGRL using the t-SNE algorithm. As it can be seen from Fig. 7, Fig. 7a is the raw features, and Fig. 7b is learned features by the SimGRL model. SimGRL’s 2D projection shows a clearer separation, which indicates that the encoder can extract more expressive node representations for downstream tasks via our proposed method.
Conclusions
In this paper, we propose a simple self-supervised graph representation learning framework, called SimGRL. Compared with the prior methods, our method achieves great performance with only one encoder and avoids the requirement of data augmentations. Using the triplet loss based on adjacency information, only one negative node is sufficient to make the model obtain significant performance, simultaneously reducing memory overhead. We conduct extensive experiments on node classification and graph classification tasks to evaluate our method. Empirical experimental results show that SimGRL has a competitive performance compared with not only unsupervised methods but also supervised methods in downstream tasks. Although our method achieved remarkable results based on the homophily assumption, there are graphs with heterophily in the real world. In future work, we intend to extend our method to be applicable in both homophilic and heterophilic settings.
Data availability
The data of this study are available from the author, [Da Huang], upon reasonable request.
References
Adhikari B, Zhang Y, Ramakrishnan N, Prakash BA (2018) Sub2vec: feature learning for subgraphs. Pacific-Asia Conference on knowledge discovery and data mining. Springer, Berlin, pp 170–182
Battaglia PW, Hamrick JB, Bapst V, Sanchez-Gonzalez A, Zambaldi V, Malinowski M, Tacchetti A, Raposo D, Santoro A, Faulkner R et al. (2018) Relational inductive biases, deep learning, and graph networks. arXiv preprint arXiv:1806.01261
Borgwardt KM, Kriegel HP (2018) Shortest-path kernels on graphs. In: Fifth IEEE international conference on data mining, pp. 74–81. IEEE
Chen T, Kornblith S, Norouzi M, Hinton G (2020) A simple framework for contrastive learning of visual representations. In: International conference on machine learning, pp. 1597–1607
Chen X, He K (2021) Exploring simple siamese representation learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 15750–15758
Defferrard M, Bresson X, Vandergheynst P (2016) Convolutional neural networks on graphs with fast localized spectral filtering. In: Proceedings of the International Conference on Neural Information Processing Systems, pp. 3844–3852
Devlin J, Chang MW, Lee K, Toutanova K (2019) Bert: Pre-training of deep bidirectional transformers for language understanding. In: NAACL-HLT
Dong X, Shen J (2018) Triplet loss in siamese network for object tracking. In: Proceedings of the European conference on computer vision, pp. 459–474
Feng Y, Wang H, Hu HR, Yu L, Wang W, Wang S (2020) Triplet distillation for deep face recognition. In: 2020 IEEE International Conference on Image Processing, pp. 808–812. IEEE
Gao T, Yao X, Chen D (2021) Simcse: Simple contrastive learning of sentence embeddings. In: Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, EMNLP 2021, Virtual Event / Punta Cana, Dominican Republic, 7-11 November, 2021, pp. 6894–6910. Association for Computational Linguistics
Grover A, Leskovec J (2016) node2vec: Scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 855–864
Hamilton WL, Ying R, Leskovec J (2017) Inductive representation learning on large graphs. In: Proceedings of the 31st International Conference on Neural Information Processing Systems, pp. 1025–1035
Hjelm RD, Fedorov A, Lavoie-Marchildon S, Grewal K, Bachman P, Trischler A, Bengio Y (2018) Learning deep representations by mutual information estimation and maximization. In: International Conference on Learning Representations
Hoffer E, Ailon N (2015) Deep metric learning using triplet network. In: International workshop on similarity-based pattern recognition, pp. 84–92. Springer
Hu W, Fey M, Zitnik M, Dong Y, Ren H, Liu B, Catasta M, Leskovec J (2020) Open graph benchmark: datasets for machine learning on graphs. Adv Neural Inf Process Syst 33:22118–22133
Huang Q, Yamada M, Tian Y, Singh D, Chang Y (2022) Graphlime: Local interpretable model explanations for graph neural networks. IEEE Trans Knowl Data Eng 2:1–6
Jin D, Yu Z, Jiao P, Pan S, He D, Wu J, Yu P, Zhang W (2021) A survey of community detection approaches: From statistical modeling to deep learning. IEEE Trans Knowl Data Eng 2:1
Kefato ZT, Girdzijauskas S (2021) Self-supervised graph neural networks without explicit negative sampling. In: The International Workshop on Self-Supervised Learning for the Web (SSL’21), at WWW’21
Kipf TN, Welling M (2016) Variational graph auto-encoders. arXiv preprint arXiv:1611.07308
Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: International Conference on Learning Representations
Kondor R, Pan H (2016) The multiscale laplacian graph kernel. In: Advances in neural information processing systems, pp. 2990–2998
Kriege N, Mutzel P (2012) Subgraph matching kernels for attributed graphs. In: International Conference on Machine Learning, pp. 291–298
Kriege NM, Giscard PL, Wilson R (2016) On valid optimal assignment kernels and applications to graph classification. In: Advances in Neural Information Processing Systems, pp. 1623–1631
Lin H, Fu Y, Lu P, Gong S, Xue X, Jiang YG (2019) Tc-net for isbir: Triplet classification network for instance-level sketch based image retrieval. In: Proceedings of the 27th ACM International Conference on Multimedia, pp. 1676–1684
Narayanan A, Chandramohan M, Venkatesan R, Chen L, Liu Y, Jaiswal S (2017) graph2vec: Learning distributed representations of graphs. arXiv preprint arXiv:1707.05005
Nikolentzos G, Siglidis G, Vazirgiannis M (2021) Graph kernels: a survey. J Artif Intell Res 72:943–1027
Pan S, Hu R, Long G, Jiang J, Yao L, Zhang, C (2018) Adversarially regularized graph autoencoder for graph embedding. In: International Joint Conference on Artificial Intelligence
Perozzi B, Al-Rfou R, Skiena S (2014) Deepwalk: Online learning of social representations. In: Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 701–710
Rhee S, Seo S, Kim S (2018) Hybrid approach of relation network and localized graph convolutional filtering for breast cancer subtype classification. In: Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, IJCAI 2018, July 13-19, 2018, Stockholm, Sweden, pp. 3527–3534
Sen P, Namata G, Bilgic M, Getoor L, Galligher B, Eliassi-Rad T (2008) Collective classification in network data. AI Mag 29(3):93–93
Shervashidze N, Schweitzer P, Van Leeuwen EJ, Mehlhorn K, Borgwardt KM (2011) Weisfeiler–lehman graph kernels. J Mach Learn Res 12(9):2539–2561
Shervashidze N, Vishwanathan S, Petri T, Mehlhorn K, Borgwardt K (2009) Efficient graphlet kernels for large graph comparison. In: Artificial intelligence and statistics, pp. 488–495. PMLR
Sun FY, Hoffman J, Verma V, Tang J (2020) Infograph: Unsupervised and semi-supervised graph-level representation learning via mutual information maximization. In: International Conference on Learning Representations
Thakoor S, Tallec C, Azar MG, Munos R, Veličković P, Valko M (2021) Bootstrapped representation learning on graphs
Thomas G, Flach P, Stefan W (2003) On graph kernels: Hardness results and efficient alternatives. In: Proceedings of the 16th Annual Conference on Computational Learning Theory and 7th Kernel Workshop, pp. 129–143
Veličković P, Cucurull G, Casanova A, Romero A, Lió P, Bengio Y (2018) Graph attention networks. In: International Conference on Learning Representations
Veličković P, Fedus W, Hamilton W.L, Lió P, Bengio Y, Hjelm RD (2019) Deep graph infomax. In: International Conference on Learning Representations, p. 4
Wang C, Liu Z (2021) Learning graph representation by aggregating subgraphs via mutual information maximization. arXiv preprint arXiv:2103.13125
Wang D, Zhang Z, Zhou J, Cui P, Fang J, Jia Q, FangY, Qi Y (2021) Temporal-aware graph neural network for credit risk prediction. In: Proceedings of the 2021 SIAM International Conference on Data Mining (SDM), pp. 702–710. SIAM
Wu F, Souza A, Zhang T, Fifty C, Yu T, Weinberger K (2019) Simplifying graph convolutional networks. In: International conference on machine learning, pp. 6861–6871. PMLR
Wu Z, Pan S, Chen F, Long G, Zhang C, Philip SY (2020) A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst 32(1):4–24
Xu K, Hu W, Leskovec J, Jegelka S (2018) How powerful are graph neural networks? In: International Conference on Learning Representations
Yanardag P, Vishwanathan S (2015) Deep graph kernels. In: Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, pp. 1365–1374
Yang Z, Cohen W, Salakhudinov R (2016) Revisiting semi-supervised learning with graph embeddings. In: International conference on machine learning, pp. 40–48. PMLR
Yao L, Mao C, Luo Y (2019) Graph convolutional networks for text classification. Proc AAAI Conf Artif Intell 33:7370–7377
You J, Ying R, Leskovec J (2019) Position-aware graph neural networks. In: International conference on machine learning, pp. 7134–7143. PMLR
You Y, Chen T, Sui Y, Chen T, Wang Z, Shen Y (2020) Graph contrastive learning with augmentations. Adv Neural Inf Process Syst 33:5812–5823
Zhao D, Chen C, Li D (2021) Multi-stage attention and center triplet loss for person re-identication. Appl Intell 2:1–13
Zhu Y, Xu Y, Yu F, Liu, Q, Wu, S, Wang, L (2020) Deep graph contrastive representation learning. arXiv preprint arXiv:2006.04131
Zhu Y, Xu Y, Yu F, Liu Q, Wu S, Wang L (2021) Graph contrastive learning with adaptive augmentation. Proc Web Conf 2021:2069–2080
Acknowledgements
This work was partly supported by the National Natural Science Foundation of China (U1701266), the Guangdong Provincial Key Laboratory Project of Intellectual Property and Big Data (2018B030322016), Special Projects for Key Fields in Higher Education of Guangdong, China (2020ZDZX3077), Guangdong Province Key Construction Discipline Scientific Research Capacity Improvement Project (2022ZDJS013), the Natural Science Foundation of Guangdong Province, China (2022A1515011146).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, D., Lei, F. & Zeng, X. SimGRL: a simple self-supervised graph representation learning framework via triplets. Complex Intell. Syst. 9, 5049–5062 (2023). https://doi.org/10.1007/s40747-023-00997-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-023-00997-6