Abstract
The success of deep learning in skin lesion classification mainly depends on the ultra-deep neural network and the significantly large training data set. Deep learning training is usually time-consuming, and large datasets with labels are hard to obtain, especially skin lesion images. Although pre-training and data augmentation can alleviate these issues, there are still some problems: (1) the data domain is not consistent, resulting in the slow convergence; and (2) low robustness to confusing skin lesions. To solve these problems, we propose an efficient structural pseudoinverse learning-based hierarchical representation learning method. Preliminary feature extraction, shallow network feature extraction and deep learning feature extraction are carried out respectively before the classification of skin lesion images. Gabor filter and pre-trained deep convolutional neural network are used for preliminary feature extraction. The structural pseudoinverse learning (S-PIL) algorithm is used to extract the shallow features. Then, S-PIL preliminarily identifies the skin lesion images that are difficult to be classified to form a new training set for deep learning feature extraction. Through the hierarchical representation learning, we analyze the features of skin lesion images layer by layer to improve the final classification. Our method not only avoid the slow convergence caused by inconsistency of data domain but also enhances the training of confusing examples. Without using additional data, our approach outperforms existing methods in the ISIC 2017 and ISIC 2018 datasets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skin cancer is a common disease that afflicts people’s health. There are many types of skin cancer, among which melanoma is a very serious cancer that can be fatal if not treated timely. However, if melanoma is diagnosed early, there is a high probability of saving the patient. Millions of skin cancer patients need to be diagnosed by dermatologists each year, and dermatologists have a lot of work to do. If computers can effectively assist dermatologists in screening skin cancer patients, then dermatologists can devote more experience to treating skin cancer patients.
The rapid development of deep learning has shown great benefits in various aspects, particularly in medical image analysis [19, 23, 24]. Recently, the important role of deep learning in skin lesion classification has been widely confirmed [3, 8, 10, 37].
In general, we anaylze that the effectiveness of deep learning technologies in skin lesion classification mainly comes from two aspects: (1) ultra-deep neural network architectures, and (2) significantly large training data sets. Firstly, cutting-edge deep neural networks, especially the convolutional neural networks that have achie-ved impressive performance in computer vision, usually have dozens of, or even hundreds of convolutional layers to extract high-level abstract visual features. Although powerful, optimizing these deep neural networks is commonly quite time-consuming, thus can probably delay the diagnosis of skin cancer, putting patients lives at risk. Secondly, larger datasets can help force deep neural networks to learn more representative patterns that are critical for the robust recognition of different skin lesions, reducing the risk of overfitting. However, since collecting lesion data from patients and annotating the collected lesion data is quite difficult, existing publicly available skin lesion datasets may not provide sufficient data to help obtain a very robust and accurate deep neural network for skin diagnosis.
To tackle above issues, researchers have proposed various improved training strategies to eithor reduce the training periods of deep neural networks or increase the diversity of training data to augment datasets. For instance, many studies apply the deep neural networks that are intensively pre-trained on ImageNet datasets for the skin lesion task [8, 27]. The pre-trained neural networks usually already have a decent representational power to describe various visual pattern, thus can promisingly accelerate the convergence speed on a new dataset. In addition, to expand the capacity of skin lesion datasets, researchers have developed different data augmentation technologies [3, 37], enriching the data examples of the original dataset automatically to help obtain a more robust skin lesion classification model.
Despite these progress, we find that current approach-es are still sub-optimal to address the long training period and limited training data issues due to the following potential reasons: (1) there is usually a significant domain gap between the pre-training datasets like the ImageNet and the skin lesion datasets. Therefore, it may still take quite a long time for current deep neural networks to converge on the new dataset; (2) current algorithms that extend skin lesion datasets do not explicitly model domainspecific information for the new dataset, which means that the extended data may lie in a different domain from the original dataset, making the optimized deep neural networks still vulnerable to some confusing skin lesions that are difficult to recognize.
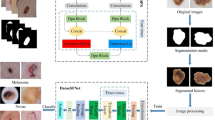
In this work, we propose a hierarchical representation learning (HRL) method, which enhances the network’s ability to learn the features of skin lesion images and to classify skin lesion images. Specifically, the proposed HRL method includes three levels of representation learning: preliminary feature extraction stage, shallow network feature extraction stage, and deep learning feature extraction stage. In the stage of preliminary feature extraction, Gabor filter is adopted to extract local texture features efficiently and pre-trained deep convolutional neural network (DCNN) provides global feature. In the stage of shallow network feature extraction, we propose a structural pseudoinverse learning algorithm (S-PIL) on the basis of pseudoinverse learning algorithm. Global image information and image edge information—Gabor features are combined as the input data of S-PIL. S-PIL is a shallow network, which has the advantage of fast training speed. It can efficiently extract the shallow feature information of skin lesion images, preliminarily identifies the skin lesion images, and distinguish the confusing images for subsequent deep learning feature extraction. In the deep learning feature extraction stage, the training data set is mixed with confusing images. It makes the deep learning network strengthen the feature learning and training of confusing skin lesion images. The whole HRL method, from simple features to complex features, analyzes skin lesion images layer by layer to improve the final classification.
In the experiments, our method outperforms other existing methods on ISIC 2017 and ISIC 2018 dataset without using additional data. In particular, the approach we proposed performs well on the area under the curve (AUC) and the overall balanced accuracy (BACC), and it has a fast convergence speed and can get the optimal result faster and better.
The main contributions of this paper are as follows:
(I) In this paper, the HRL method is proposed for the classification of skin lesion images. The proposed method uses three levels of feature learning, namely preliminary feature extraction, shallow feature extraction and deep learning feature extraction. HRL method analyzes the features of skin lesion images layer by layer from simple features to complex features to improve the final classification.
(II) The S-PIL algorithm proposed in this paper is an efficient shallow network, whose input layer combines the global information of the image and the local texture information—Gabor features. It makes further feature extraction on skin lesion images, and can efficiently identifies confusing skin lesion images for the subsequent deep learning feature extraction.
(III) The proposed method mix the confusing skin lesion images into the original data set, change the domain features of the training set, and enhance the training of confusing images by deep learning network. Our method can outperform other existing methods without using additional data.
The rest of this paper is organized as follows. In second section, the related works are presented, including skin lesion classification, pseudoinverse learning algorithm and Gabor filter. In third section, the proposed method is introduced in details. Fourth section is experimental results and analysis that show the datasets, model training, experimental results and performance analysis. Fifth section is discussion about the advantage of the proposed method and the future work. Sixth section is conclusion about this paper.
Related work
Skin lesion classification
Automatic skin cancer detection has been studied for decades. Before the advent of deep learning, many methods adopted manually designed features and then applied typical machine learning algorithms to recognize skin lesions based on extracted features [22]. Instead of empirical features, the rapid development of deep neural networks and deep learning has helped researchers to develop highly accurate skin recognition models based on deep visual features. Deep learning technologies are increasingly important in medical image processing. For example, in the ISIC, an important competition for skin cancer image analysis, the teams that achieved good results almost all adopted deep learning methods. For example, some methods use deep learning to extract high-level features and then use these features to perform classification [28]. Currently, the more commonly used method is the end-to-end deep convolutional neural networks with transfer learning. In particular, Esteva et al. [8] studied the utilization of pre-trained deep neural network architectures [33] for the end-to-end skin cancer classification, and a big dataset (129,450 clinical images) was used in their study. They achieved promising classification results which are comparable to those of human dermatologists. In addition, there are other ways to use deep learning to detect skin cancer. It will improve the classification effect that segmentation is carried out before classification to obtain the areas of skin lesion [27]. Besides using separate network, some works use ensemble learning to integrate the results of multiple deep learning models for better results [4, 28, 29]. There is also work that combines image data with other forms of data, such as medical record information, to classify skin diseases using deep learning [31].
Although the popular deep learning has achieved good results in the task of skin lesion detection, it still has the problems of long training time and limited training data. Moreover, the existing skin lesion detection methods do not model the intra-class variations and inter-class similarities, and some of them have limited discrimination ability for the confusing skin lesion images.
Pseudoinverse learning algorithm
Pseudoinverse learning algorithm (PIL) [12,13,14] was first proposed by Guo et al., which is a fast algorithm to train feedforward neural network. The basic idea is to find a set of orthogonal vector bases, make the output vectors of hidden layer neurons tend to be orthogonal by using nonlinear activation function, and then approximate the output weights of the network by calculating the pseudoinverse. The PIL algorithm is simple and easy to use. It only requires efficient matrix inner product and pseudoinverse operations. Moreover, different from the neural networks which mainly optimized by stochastic gradient descent (SGD) according to minibatches, the PIL algorithm can approximate the optimal solution across all the dataset , which is crucial for stable and balanced training.
The principle of PIL algorithm is as follows:
Suppose that the training data set \(D = \{ X^i, O^i\}_{i=1}^n\) denotes n samples, where \(X^i = {(x_1, x_2,...,x_d)} \in R^d\) and \(O^i = {(o_1, o_2,..., o_m)}\in R^m\) denotes i-th input sample and the corresponding expected output respectively. O is the output data, which is given in advance. For a single hidden layer forward network, the most used sum-of-square objective function is as follows,
where \(g_j (x^i,\theta )\) is j-th output neuron, which shows the map from input value into predicted value, and it is defined as follows,
where \(\theta \) is the parameters of entire network, q is the number of hidden neurons,\( w_{k,l}^0\) is the input weight between k-th input neuron and lth hidden neuron, \(w_{l,j}^1\) is the output weight between l-th hidden neuron and j-th output neuron, b is bias. For better understanding, the above formula is written as matrix form,
where \(W^1\) is the output weight, H is the hidden output, which is written as follows,
where \(W^0\) is the input weight, which can be obtained by random selecting from uniform distribution. We can get the output weight,
where \(H^+\) is the pseudoinverse matrix of H. \(H^+\) can be calculated by Moore-Penrose inverse, then the output weight is,
where \(\lambda \) is a regularization parameter. However, it is difficult to seamlessly apply the original PIL algorithm to learn features in lesion data. First, the original PIL algorithm only learn patterns from the global information of input images and thus the detailed local patterns are not explicitly utilized to minimize the objective. Since it is more important to identify the edge patterns on lesion areas, the original PIL would not be effective to recognize hard patterns in lesion data. Second, it is impractical to apply the original PIL algorithm on ultra-deep neural networks. By increasing the depth of a neural network significantly, the pseudoinverse should be computed recursively, which is usually intractable. Without the deep abstraction of visual patterns, the resulting model will only have limited expressive capacity to encode information, especially the hard patterns.
Gabor filter
Gabor filter is a linear filter used for edge extraction. Its frequency and direction expression is similar to that of the human visual system, which can provide good directional selection and scale selection characteristics. Gabor is not sensitive to light changes, so it is very suitable for texture analysis [7]. As an handcraft feature, Gabor feature is easy to be extracted quickly. The extraction process of features with Gabor filter is as follows:
where I is the grayscale distribution of the image, \(I_G\) is the feature extracted from I, \( \oplus \) stands for 2D convolution operator, G is the defined Gabor filter.
The two-dimensional Gabor kernel function is defined as follows [26]:
Eq. 8 is obtained by the multiplication of a Gaussian function and a cosine function. The arguments x and y specify the position of a light impulse, where \((x_0, y_0)\) is the center of the receptive field in the spatial domain. \(\theta \) is the orientation of parallel bands in the kernel of Gabor filter, and the valid values are real numbers from 0 to 360. \(\varphi \) is the phase parameter of cosine function in Gabor kernel function, and the valid values is from \(-\,\)180 to 180\(^\circ \). \(\gamma \) is the space aspect ratio, which represents the ellipticity of the Gabor filter. \(\lambda \) is the wavelength parameter of the cosine function in the Gabor kernel function. \(\sigma \) is the standard deviation of Gaussian function in the Gabor kernel function. This parameter determines the size of acceptable area in the Gabor filter.
Representation learning
Representation learning generally refers to the process of automatically learning effective data features and it is divided into unsupervised representation learning and supervised representation learning [2]. Unsupervised representation learning includes autoencoder, clustering algorithm [16, 21], etc. The most popular form of supervised representation learning is deep learning. DCNN has strong representational learning ability. It can extract features from input data according to layer structure, obtain effective feature representation, and use effective features to perform data classification, regression and other operations [18]. In 2012, AlexNet [25] was proposed. Since then, with the development of computer hardware and the improvement of computing power, many effective deep learning models have emerged successively, such as GoogleNet [32], ResNet [17], DenseNet [20] and EfficientNet [34]. Generally, DCNNs require a large amount of training data and excessive computational resources, and the training is time-consuming. However, for skin cancer image processing, a large amount of labeled training data is difficult to obtain.
Proposed method
To address the issues of long training periods and poor robustness to confusing examples in the classification task of skin lesion images, we propose the efficient structural pseudoinverse learning-based hierarchical representation learning for skin lesion classification. Since there are slight differences among different classes in the skin lesion image data, it is very important to obtain knowledge from the training data to improve the result of classification.
As shown in Fig. 1, the proposed HRL method includes three levels of feature extraction: preliminary features extraction, shallow network features extraction and deep learning features extraction. The three levels of representation learning, from simple features to complex features, analyze skin lesion images layer by layer to obtain knowledge for subsequent classification tasks. Specifically, the preliminary features consist of two parts: the global feature \(X_\mathrm{g}\) obtained by the pre-trained DCNN and the handcraft features \(X_\mathrm{l}\) extracted by Gabor filter. These two features correspond to the global feature and local texture features of the skin lesion image respectively. The preliminary features can be obtained efficiently, making preparation for the subsequent shallow network feature learning. The shallow network features extraction is realized by S-PIL algorithm, which is based on PIL algorithm. Unlike PIL, S-PIL’s input layer combines the global feature \(X_\mathrm{g}\) with the local texture feature \(X_\mathrm{l}\). S-PIL can efficiently analyze skin lesion information and identify examples that are hard to classify. The confusing examples are used to prepare for the deep learning features extraction. In the deep learning features extraction stage, different from the traditional DCNN, we increase the proportion of confusing examples in the training data set to enhance the learning of DCNN on confusing examples. It can effectively improve the classification results of skin lesion images by focusing on the confusing examples.
Shallow neural network and deep CNN are trained separately. We first train shallow neural network to find confusing examples, and then train the deep CNN. Shallow neural network is trained as described in “Shallow network features” section, and deep CNN is trained as described in “Deep learning features” section.
The proposed HRL method is an effective strategy to improve the classification result of skin lesions. The three levels of representation learning analyze skin lesion images layer by layer to learn knowledge from the data. HRL method can improve the classification result of skin lesion images, and this method is very efficient to implement. The detailed descriptions of the HRL method will be shown in the following sections.
Preliminary features
In our proposed method, the preliminary features contain global feature and local texture features. The global feature is based on the feature extracted using the pre-trained DCNN. It provides high-level abstract semantics learned from pre-trained datasets. In practice, we consider the output of the last pooling layer of the Resnet50 pre-trained on Imagenet as the global feature \(I_g\). For local features, we use Gabor filter to extract local texture features. The difference between skin lesion images is very subtle, so Gabor features are used to represent the texture characteristics of skin lesion images. We use \(I_l\) to denote the local Gabor features. In practice, the parameters of Gabor filter are as follows: the scale of the Gabor filter is 7, the wave length \(\lambda \) is \(\pi /2\), the orientation \(\theta \) is set as 8 with different orientations from 0 to \(7\pi /8\), and the difference between the two adjacent orientations is \(\pi /8\), standard deviation of the Gaussian function \(\sigma \) is 1.0, the spatial aspect ratio \(\gamma \) is 0.5, and the phase parameter \(\varphi \) is 0.
The global feature and the local Gabor features are unified to construct the structural representation \(I_\mathrm{{S}}\) for the S-PIL algorithm. The unification of each pair of the global feature and local Gabor features can be described as a data sample for learning. Formally, we define a data sample for training S-PIL as:
where + represents the unification and \(I_\mathrm{{S}}\) is a data sample for training S-PIL. In this study, we implement the + using concatenation operation, maintaining both the global feature and local Gabor features during learning.
Shallow network features
In the stage of shallow network features extraction, S-PIL is used to quickly extract shallow network knowledge. The S-PIL takes advantage of the fast convergence speed of PIL framework and quickly learns the data statistics of the skin lesion dataset, delivering a roughly accurate and indicative analysis for the skin lesion data based on the pre-trained DCNN and handcraft Gabor features. Different from the existing PIL algorithms that usually only learn from the raw input data, the proposed S-PIL effectively exploits the structural representation of skin lesion data to achieve detailed perception of the skin lesion image. The learned knowledge about the detailed understanding of skin lesion data can then adequately provide a robust and accurate estimation of confusing skin lesion image, facilitating the later training of the DCNN on the skin lesion dataset.
In particular, the structural representation adopted in the S-PIL consists of two parts, i.e. global representation and local Gabor texture representation. With the structural data obtained based on Eq. 9, we can apply the PIL framework as described in Eqs. 4–6 to implement the S-PIL:
where \(W_\mathrm{{S}}^1\) is the weight matrix to be learned from the data, \(W_\mathrm{{S}}^0\) is the randomly sampled input weights used in the PIL, and f is the squashing function. According to the PIL framework, we can efficiently obtain \(W_\mathrm{{S}}^1\) by optimizing the following objective:
Then, we have:
Based on the results of S-PIL, we can obtain a roughly accurate estimation of the skin lesion data efficiently. Such preliminary knowledge can facilitate us to identify the more confusing examples in the skin lesion dataset. The loss of each skin lesion image obtained by S-PIL is \(\mathrm{{Loss}}_{I_\mathrm{{S}}}^i\), which can be used to determine the confusing examples. Afterwards, we rank the skin lesion data examples in a descending order according to the loss values of image across the dataset. We identify the confusing examples by taking top-10% of the sorted skin lesion data. By increasing the importance of the confusing examples, we can help DCNNs achieve better training performance.
S-PIL is an efficacious prior-training domain analyzing mechanism to extract adequate knowledge that can help migrate domain statistics from the pre-trained data distribution to the skin lesion data distribution. S-PIL identifies the confusing examples from the learned prior-training domain knowledge to improve the convergence of DCNNs on the skin lesion datasets which only have a limited amount of data examples for training.
Deep learning features
In the deep learning features extraction stage, the main idea of the proposed method is to increase the proportion of confusing examples identified by S-PIL in the training set, so as to improve the training of DCNN, make it converge faster, and improve the robustness of the confusing examples.
Mathematically, we use \(X_\mathrm{L}\) to represent the set of lesion data examples. \(f_\mathrm{{cls}}(W,X_\mathrm{L})\) is used to refer the the classification function achieved by a DCNN. Currently, the parameters of DCNN classification models, denoted as W, are generally pre-trained using external datasets such as ImageNet which are usually from a very different data distribution from the lesion data \(X_\mathrm{L}\).
We denote the pre-trained parameters as \(W^\mathrm{{pre}}\). In general, the typical training procedure is defined to minimize the following objective:
where L is the overall loss function, Y is the set of ground-truth labels corresponding to the data examples from \(X_\mathrm{L}\), and \(f_\mathrm{{cls}}\) is the classification function implemented by a DCNN.
Different from current training strategy that directly fine-tunes W from \(W^\mathrm{{pre}}\), we propose to first build an efficient learning function, denoted as \(f_X\), to analyze the characteristics of the new skin lesion data domain based on \(W^\mathrm{{pre}}\) before training the DCNN parameters W. Then, we attempt to identify confusing examples based on the learned knowledge of the skin lesion data domain and use these confusing examples to adequately modifying the domain statistics of \(X_\mathrm{L}\), effectively improving the optimization of W. Therefore, the overall objective of our training strategy is:
where \(X^N_{L}\) is the modified data distribution from \(X_\mathrm{L}\) based on the knowledge learned by \(f_X\):
where \(f_\mathrm{{{S{-}PIL}}}\) represents the efficient S-PIL algorithm. In this study, we mainly implement \(f_X\) by re-weighting the importance of confusing examples identified based on the S-PIL algorithm.
Most of the existing methods classify skin lesion images by using DCNNs pre-trained from natural images, then learn a limited number of skin lesion images \(X_\mathrm{L}\), and finally obtain a network model. Unlike existing methods, we used pre-trained DCNNs to learn the original skin lesion data and the confusing examples we found. Specifically, we mix the confusing examples with the original skin lesion images. During the training of DCNNs, we redistributed the weight of the original skin lesion images and the confusing examples. We obtain the new dataset \(X_\mathrm{L}^N\) , which changes the data distribution of the original data set and increases the frequency of confusing examples. Through experimental analysis, we find that the effect is better when the weight ratio between the original skin lesion image and the confusing example is 1:2. The new dataset \(X_\mathrm{L}^N\) not only preserves most skin lesion images, but also increases the proportion of confusing examples. In other words, we can learn all the information contained in skin lesion images and strengthen the learning of confusing examples.
Experimental results and analysis
Datasets
The ISIC 2017 dataset [6] and ISIC 2018 dataset [5, 35] are used to estimate our method. As shown in Table 1, ISIC 2017 dataset includes 3 classes of dermatological images: Melanoma, Seborrheic keratosis and Nevus. It consists of 2750 images, which are divided into three datasets: training set (2000 images), validation set (150 images), and test set (600 images). We conducted ablation experiments on ISIC 2017 dataset to demonstrate the effectiveness of our approach, and we also compared our method with other baseline approaches on ISIC 2017. We do not use external training data on the experiments.
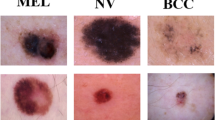
We also carried out experiments on the ISIC 2018 dataset to compare it with existing state of the art methods. ISIC 2018 dataset is more complex than ISIC 2017 dataset. As shown in Table 2, ISIC 2018 includes 7 classes of skin lesion images: Melanoma, Nevus, Basal hominins cell carcinoma, Actinic keratosis, Benign keratosis, Dermatofibroma, Vascular lesion. ISIC 2018 data-set includes 10,015 train data, 193 validation data, 1512 test data. However, validation data and test data have no ground-truth labels. We randomly selected training data (80%) and validation data (20%) from train dataset. Similar to the previous experiment, we do not use external training data. The results are uploaded to the ISIC online platform for evaluation. Both ISIC 2017 and ISIC 2018 datasets are imbalanced, with Nevus having the largest amount of data.
Model training
The pre-trained DCNNs we chose to use for the final classification of skin lesion images included: Resnet152, Densenet161, Resnet50, InceptionV4. We trained the network with Stochastic Gradient Descent (SGD) with a momentum factor 0.9, batch size 32, starting learning rate 1e-3. The training data is shuffled before each epoch. We also adopted data augmentation, such as color conversion, random crops, rotate the image by up to 90, randomly flip the images horizontally and vertically. Each model was trained for at most 100 epochs, with an early stopping criterion. For the ISIC 2017 data set, we resize the image into 1024 before experiment. Our experiments were conducted on NVIDIA Titan X.
We test our method on the test dataset, and the evaluation criteria for experimental results are mainly derived from the confusion matrix. Specifically, we evaluate our model on ISIC 2017 dataset according to the area under the curve (AUC), accuracy (ACC), sensitivity (SE), specificity (SP). And the ISIC 2018 dataset is evaluated by the overall balanced multi-class accuracy (BACC), AUC, ACC.
Results on ISIC 2017
We compared our method with other existing methods, and the results are shown in Table 3. The methods listed in Table 3 are from the ISIC 2017 challenge and existing papers using ISIC 2017 data. In our method, the first 200 examples with the largest loss are selected as the confusing examples. The ratio of the original examples to the confusing examples is 1:2. Resnet152 was used to train up to 50 epochs, with early stop mechanism.
In ISIC 2017, the main indistinct classes are Melanoma and Seborrheic keratosis, in which Melanoma is a skin disease threatening human life and health. Table 3 shows the classification results of Melanoma and Seborrheic keratosis, as well as the average results. It also shows whether these methods use external training data. Figure 2 is the normalized confusion matrixes of different methods on ISIC 2017 testing set. MEL, SK, NV are abbreviations of Melanoma, Seborrheic keratosis, Nevus respectively. The results show that the proposed method can get a better classification result without using external training data. Our method performs better in AUC and ACC than other methods and balances the performance of SE and SP. Specifically, our method improves the mean AUC and mean ACC by at least 1.2% and 3.5% compared with other existing methods.
Results on ISIC 2018
We also conducted experiments on ISIC 2018, and compared the experimental results with those state of the art methods. The methods in Tables 4 and 5 are mainly from the top methods in ISIC 2018 challenge. R stands for ranking, such as, R1 is the first place in ISIC 2018 challenge. The ISIC 2018 data is more complex and the category imbalance is more severe, so we added BACC evaluation metric when comparing the results. We did not use external training data, and used Senet154 to train 100 epochs with early stopping mechanism.
Table 4 shows the results on different methods without using extra data. Our approach has advantages in the performance of AUC and BACC. R3 is the third place method in ISIC 2018, and it is the first place in methods without using extra data in ISIC 2018 challenge. Our approach performs better than R3 in AUC and BACC. Specifically, our method improves the AUC by 0.5% and the BACC by 1.2% compared with R3 method.
Table 5 shows the results on different methods using extra data. The italic numbers in the table indicate the second place among these methods. R1 and R2 is the first place and second place methods in ISIC 2018 challenge, and they all use extra data. Our method performs comparable to the R1 and R2 method without using additional data.
Performance analysis
The effectiveness of HRL
In order to prove the effectiveness of our method, we test Resnet50, Resnet152, InceptionV4, Densenet161, and the corresponding CNN adding our method. In the ISIC 2017 dataset, Melanoma and Keratosis are the most difficult to be classified, and we present classification results of them. The experimental results are shown in Fig. 3. Our method significantly improves CNN’s performance in classifying skin lesions.
Figure 4 shows the confusion matrix for Densenet161 and Densenet161-HRL on the ISIC 2018 validation dataset. ISIC 2018 dataset is randomly divided into train dataset (80%) and Validation dataset (20%). Densenet161 and Densenet161-HRL are trained respectively to obtain the confusion matrix on the validation set. From Fig. 4, we can see that our method performs better and can classify more samples correctly.
We evaluate the convergence of our proposed method on ISIC 2017 data set. We compared the convergence and convergence speed of the Resnet152 and Resnet152 with HRL strategy. As shown in Figs. 5 and 6, both methods can converge, but our proposed method can converge faster. On ISIC 2017 train dataset, Resnet-152 with HRL converges around the 15th epoch and Resnet152 converges around the 20th epoch. On ISIC 2017 test dataset, Resnet152 with HRL converges around the 10th epoch and Resnet152 converges around the 15th epoch.
Ablation experiments
Table 6 shows the results of the ablation experiments on ISIC 2017 data set, which is the classification of Melanoma on the test set. Resnet152 was used in deep learning feature. In the first experiment, training data was directly fed into PIL without the preliminary feature, and confusing examples were found, then DCNN was trained. In the second experiment, the preliminary feature is directly input into DCNN for training. In the third experiment, the classification results were obtained by using preliminary feature to train S-PIL. In the fourth experiment, Gabor feature is not used in preliminary feature. The fifth experiment used the complete HRL method. The results in Table 6 show that all three parts of the HRL method are indispensable. Without deep learning feature and without shallow network feature have a great impact on the experimental results. Specifically, AUC is reduced by 7% and 3.3% compared with HRL respectively. Without preliminary feature and without Gabor feature also affected the experimental results. Specifically, AUC was 1.4% and 1.1% lower than HRL respectively.
Discussion
Deep learning has shown remarkable success in automatically identifying skin lesion images. The success of deep learning mainly depends on a large number of correctly labeled samples and super-deep networks. However, large numbers of labeled samples are difficult to obtain, especially medical images. Although techniques such as transfer learning and data augmentation can alleviate the problem of lack of data, deep learning still has disadvantages such as slow convergence and low robustness to confusing skin lesion images.
In order to solve the above problems, we think it is important to learn enough knowledge of skin lesion images from samples for classification. Therefore, we propose the HRL method to learn the features of skin lesion images layer by layer. Traditionally, the transfer learning is directly applied to the classification of skin lesion images, without considering the serious inconsistency in the data domain, and just uses brute-force methods to learn statistical knowledge from a large number of data. Our HRL method fully learns the feature knowledge before the transfer learning, which makes the deep network focus more on the learning of the confusing examples and improve the convergence speed of the network, which is conducive to obtain the better performance in the classification of skin lesion images. The HRL method we designed has achieved good results on ISIC 2017 and ISIC 2018 datasets.
Though the HRL method achieves good results in the classification of skin lesions, the current HRL method is not end to end, the early stage still needs to be designed artificially, and the quality of the design has an impact on the subsequent classification. S-PIL is used in HRL to take the advantage of fast training of PIL algorithm so as to obtain preliminary classification information. In fact, other simple networks could be used, but they may not be as fast as S-PIL. The HRL method proposed in this paper is a scheme to improve the automatic classification of skin lesions. As mentioned above, while HRL does work well, there are still drawbacks, such as not being end-to-end, not trying more shallow networks, etc. We will optimize the HRL method in the future.
Conclusion
This paper proposes the structural pseudoinverse learning-based hierarchical representation learning method to efficiently classify skin lesions. HRL method includes three levels of feature extraction. The first layer is preliminary feature extraction. Global features from pre-trained DCNN and local features from handcrafted feature Gabor are adopted to make a comprehensive and detailed analysis of skin lesion images. The second layer is shallow network feature extraction. The S-PIL uses structural representation input to quickly identify skin lesion images and screen out confusing samples for subsequent deep learning feature extraction. The third layer is deep learning feature extraction stage. Different from the traditional classification method using pre-trained DCNN, we increased the proportion of confusing samples in training data. Our method can relieve the slow convergence caused by inconsistent data domain in the pre-training method, and solve the problem that DCNNs need large training data, improve the robustness of the network. We evaluates our method on the ISIC 2017 and ISIC 2018 datasets without additional data, and the results show that our method performs better than other existing methods.
Change history
31 January 2022
A Correction to this paper has been published: https://doi.org/10.1007/s40747-022-00654-4
References
Barata C, Marques JS, Celebi ME (2019) Deep attention model for the hierarchical diagnosis of skin lesions. In: IEEE Conference on Computer Vision and Pattern Recognition Workshops, CVPR Workshops 2019, Long Beach, CA, USA, June 16-20, 2019, Computer Vision Foundation / IEEE, pp 2757–2765
Bengio Y, Courville A, Vincent P (2013) Representation learning: A review and new perspectives. IEEE transactions on pattern analysis and machine intelligence 35(8):1798–1828
Bissoto A, Perez F, Valle E, Avila S (2018) Skin lesion synthesis with generative adversarial networks. In: OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis, Springer, pp 294–302
Celebi Emre M, Codella Noel, Halpern Allan (2019) Dermoscopy image analysis: Overview and future directions. IEEE Journal of Biomedical and Health Informatics 23(2):474–478
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti Ma (2019) Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). https://arxivorg/abs/190203368
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, et al. (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, pp 168–172
Daugman J (1980) Two-dimensional spectral analysis of cortical receptive field profiles. Vision Research 20(10):847–856
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115
Gessert N, Sentker T, Madesta F, Schmitz R, Kniep H, Baltruschat I, Werner R, Schlaefer A (2018) Skin lesion diagnosis using ensembles, unscaled multi-crop evaluation and loss weighting. http://arxivorg/abs/180801694
Gessert N, Sentker T, Madesta F, Schmitz R, Kniep H, Baltruschat I, Werner R, Schlaefer A (2020) Skin lesion classification using cnns with patch-based attention and diagnosis-guided loss weighting. IEEE Transactions on Biomedical Engineering 67(2):495–503
González-Díaz I (2019) Dermaknet: Incorporating the knowledge of dermatologists to convolutional neural networks for skin lesion diagnosis. IEEE Journal of Biomedical and Health Informatics 23(2):547–559
Guo P, Lyu MR (2004) A pseudoinverse learning algorithm for feedforward neural networks with stacked generalization applications to software reliability growth data. Neurocomputing 56:101–121
Guo P, Chen CP, Sun Y (1995) An exact supervised learning for a three-layer supervised neural network. In: Proceedings of 1995 International Conference on Neural Information Processing, pp 1041–1044
Guo P, Lyu MR, Mastorakis N (2001) Pseudoinverse learning algorithm for feedforward neural networks. Advances in Neural Networks and Applications pp 321–326
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. Journal of Biomedical Informatics 86:25–32
Hartigan JA, Wong MA (1979) Algorithm as 136: A k-means clustering algorithm. Journal of the royal statistical society series c (applied statistics) 28(1):100–108
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hinton GE, Salakhutdinov RR (2006) Reducing the dimensionality of data with neural networks. science 313(5786):504–507
Hu Z, Tang J, Wang Z, Zhang K, Zhang L, Sun Q (2018) Deep learning for image-based cancer detection and diagnosis- a survey. Pattern Recognition 83:134–149
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Jain AK (2010) Data clustering: 50 years beyond k-means. Pattern recognition letters 31(8):651–666
Jain S, Pise N et al (2015) Computer aided melanoma skin cancer detection using image processing. Procedia Computer Science 48:735–740
Karar ME, ezzeldinhemdan, Shouman MA (2020) Cascaded deep learning classifiers for computer-aided diagnosis of covid-19 and pneumonia diseases in x-ray scans. Complex and Intelligent Systems (8)
Kollias D, Tagaris A, Stafylopatis A, Kollias S, Tagaris G (2018) Deep neural architectures for prediction in healthcare. Complex and Intelligent Systems 4:119–131
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems 25:1097–1105
Kruizinga P, Petkov N (1999) Nonlinear operator for oriented texture. IEEE Transactions on Image Processing 8(10):1395–1407
Li Y, Shen L (2018) Skin lesion analysis towards melanoma detection using deep learning network. Sensors 18(2):556
Majtner T, Yildirim-Yayilgan S, Hardeberg JY (2016) Combining deep learning and hand-crafted features for skin lesion classification. 2016 Sixth International Conference on Image Processing Theory. Tools and Applications (IPTA), IEEE, pp 1–6
Maragoudakis M, Maglogiannis I (2010) Skin lesion diagnosis from images using novel ensemble classification techniques. In: proceedings of the 10th IEEE International Conference on Information Technology and Applications in Biomedicine, IEEE, pp 1–5
Nozdryn-Plotnicki A, Yap J, Yolland W (2018) Ensembling convolutional neural networks for skin cancer classification. International Skin Imaging Collaboration (ISIC) Challenge on Skin Image Analysis for Melanoma Detection MICCAI
Roffman D, Hart G, Girardi M, Ko CJ, Deng J (2018) Predicting non-melanoma skin cancer via a multi-parameterized artificial neural network. Scientific reports 8(1):1701
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, PMLR, pp 6105–6114
Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data 5:180161
Valle E, Fornaciali M, Menegola A, Tavares J, Avila S (2020) Data, depth, and design: Learning reliable models for skin lesion analysis. Neurocomputing 383:303–313
Yu L, Chen H, Dou Q, Qin J, Heng PA (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Transactions on Medical Imaging PP(99):994–1004
Zhang J, Xie Y, Xia Y, Shen C (2019) Attention residual learning for skin lesion classification. IEEE Transactions on Medical Imaging 38(9):2092–2103
Zhuang J, Li W, Manivannan S, Wang R, Zhang JJG, Pan J, Jiang G, Yin Z (2018) Skin lesion analysis towards melanoma detection using deep neural network ensemble. ISIC Challenge 2018:2
Acknowledgements
The research work described in this paper was fully supported by the National Key Research and Development Program of China (no. 2018AAA0100203), the Joint Research Fund in Astronomy (U2031136) under cooperative agreement between the NSFC and CAS, and the National Key Research and Development Program (no. 2017YFC 1502505). We thank Dr. Zhe Chen for his suggestions about this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Corresponding authors declare on behalf of all authors that there is no conflict of interest. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to error in text.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deng, X., Yin, Q. & Guo, P. Efficient structural pseudoinverse learning-based hierarchical representation learning for skin lesion classification. Complex Intell. Syst. 8, 1445–1457 (2022). https://doi.org/10.1007/s40747-021-00588-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00588-3