Abstract
Autism spectrum disorder (ASD) is a neurological and developmental disorder that begins early in childhood and lasts throughout a person’s life. Autism is influenced by both genetic and environmental factors. Lack of social interaction, communication problems, and a limited range of behaviors and interests are possible characteristics of autism in children, alongside other symptoms. Electroencephalograms provide useful information about changes in brain activity and hence are efficaciously used for diagnosis of neurological disease. Eighteen nonlinear features were extracted from EEG signals of 40 children with a diagnosis of autism spectrum disorder and 37 children with no diagnosis of neuro developmental disorder children. Feature selection was performed using Student’s t test, and Marginal Fisher Analysis was employed for data reduction. The features were ranked according to Student’s t test. The three most significant features were used to develop the autism index, while the ranked feature set was input to SVM polynomials 1, 2, and 3 for classification. The SVM polynomial 2 yielded the highest classification accuracy of 98.70% with 20 features. The developed classification system is likely to aid healthcare professionals as a diagnostic tool to detect autism. With more data, in our future work, we intend to employ deep learning models and to explore a cloud-based detection system for the detection of autism. Our study is novel, as we have analyzed all nonlinear features, and we are one of the first groups to have uniquely developed an autism (ASD) index using the extracted features.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD), also commonly known as autism, is a complex neurological condition. ASD is characterized by a shortfall in social behaviors and nonverbal communications, such as avoiding eye contact or facing difficulties with controlling emotions and understanding others’ emotions, in the first three years of human life [1]. Nonspecific symptoms such as abnormal sensory perception skills and experiences, inept motor skills, and insomnia are common in some children with ASD. Other symptoms include cognitive impairment, hyperactivity, and aggressive and repetitive behaviors. ASD is known to be multifactorial, stemming from both genetic and environmental influences [2]. Gene and chromosomal defects exist in approximately 10% to 20% of individuals with ASD [3], with siblings born into ASD families having a 50 times larger risk of ASD, amid a relapse rate of 5%–8% [4].
Environmental influences during prenatal, perinatal, and post-natal period also contribute to ASD [5]. Pre-natal factors such as exposure to teratogens that cause malformation of embryo[6], perinatal factors such as unusually short gestation duration or birth asphyxia, and post-natal factors such as viral infection or mercury toxicity have been attributed as possible factors contributing to ASD [7, 8]. Figure 1 presents the neural connections existing in a normal versus ASD brain. The brain of a child with autism is characterized by a surplus of connections between brain cells as compared to the brain of a normal child. This occurs due to a decline in the natural process of removing damaged or degraded neurons during brain development, causing nonuniformity in neural pattern, disconnectedness, and lack of correlation of cognitive function between different regions, affecting the brain network [9]. Hence, more dense neural connections are observed in an autistic brain [10] and thus, this explains the shortfalls in social behavior and cognitive functions in ASD children as compared to normal ones. Thus, studying brain signals and understanding the neural patterns could shed light on why autistic children exhibit certain characteristics.
With 1 in 160 children expected to suffer from ASD globally, and the occurrence of ASD rising progressively, timely intervention, and prognosis during early childhood is important [11]. ASD is typically diagnosed based on clinical assessments such as screening tests, to detect the presence of core symptoms such as lack of social interactions, communication problems, and constrained behaviors and interests in children. However, experts who may not have seen a large number of children with ASD may over- or underdiagnose [12], due to the varying diagnostic representations these children may present. This warrants the need for an automated diagnostic tool to aid clinicians in diagnosing ASD accurately. Electroencephalograms (EEG) record the brain activity, and provide useful information about the condition of the brain. Hence, brain signals are prevalently employed to study neurological disorders such as Parkinson’s disease [13], depression [14], epilepsy [15], autism [16,17,18], schizophrenia [19], attention-deficit hyperactivity disorder [20], and Alzheimer’s disease [21]. Since EEG signals are nonstationary in nature, nonlinear features are used to classify normal and abnormal EEG signals [22]. Therefore, in this work, we aim to develop a classification system for the detection of autism. This system would be able to rapidly classify EEG signals into autism versus normal. The system is developed by first extracting nonlinear features from EEG signals. Optimal features are selected with Student’s t test. The optimal feature set is then reduced using Marginal Fisher Analysis (MFA) and ranked again using the t test. The best-performing features are classified using the Support Vector Machine (SVM) with polynomials 1, 2, and 3 thereafter. Additionally, we are one of the first groups to have developed an ASD index to discern between the two classes using the three most significant features.
The remainder of this paper is organized as follows: "Introduction" and "Methodology" discuss the background of autism, and summarize studies using computer-aided detection (CAD) systems to detect autism, respectively. "Results" describes the methodology employed, and "Discussion" and "Conclusion" discuss the results obtained and compare it against other studies. Sections 7 and 8 highlight future work and conclude the paper.
Methodology
Data used
The EEG signals used in this study were obtained from 40 ASD and 37 normal children, with an age range of 4–13 years. Children with ASD were recruited from the centers of special education and from normal schools, based in Jordan. The children with ASD had been pre-diagnosed by experts of behavioral assessment. Children suffering from other neurological disorders and those on medications were excluded from the study. A parent of each child provided informed consent prior to the study.
Study protocol and EEG recording
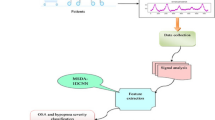
This study had been approved by the ethical committee of the Institutional Review Board (IRB) of Jordan University of Science and Technology (JUST) and King Abdullah Hospital (KAAH). EEG signals were collected from the participants (ASD and normal children) as they were seated, in the resting state, with eyes opened. The EEG specialist placed an EEG cap (64 channels ANT neuro), with the suitable size, on the head of participants. The participants were between the 3rd and 6th grades of elementary school, having an age range of 9–12 years. Normal children were recruited from regular schools (not special education schools). They were selected based on the school medical and social records, wherein they appeared with normal attitude, without any problems in school scores or health. ASD children were recruited from special education schools. All children were pre-assessed by behavior experts who diagnosed the state and severity of the child based on the different internationally established scaling methods. All children with comorbidities (based on the medical record) were excluded from this study. Twenty minutes of brain signals were recorded. The EEG cap was fixed on the participants’ scalp, following the 10–20 international standards. The affixed cap was linked through a cable to the 64-EEGO amplifier, and a desktop computer with MATLAB, EEGO software, and LA-106 ERP software. The signal at each channel was sampled at 500 Hz, and the frequency range considered was from 0.3 to 40 Hz. All EEG signals were segmented into sample lengths of 5519 data points. Figure 2 presents the proposed methodology.
Feature extraction
Eighteen nonlinear, highly distinctive features were extracted from the signals. They are the: 1. approximate entropy [23], 2. permutation entropy [24], 3. sample entropy [25], 4. Tsallis entropy [26], 5. fuzzy entropy [27], 6. Kolmogorov–Sinai entropy [28], 7. Kolmogorov complexity [29], 8. modified multiscale entropy [30], 9. Rényi entropy [31], [41], 10. Shannon entropy [32], 11. wavelet entropy [33], 12. signal activity [34], 13. Bispectrum [35], 14. Cumulant [36], 15. Hjorth [37], 16. RQA [38], 17. largest Lyapunov exponent [39], and 18. correlation dimension [40] features.
Approximate entropy is helpful to compute the complexity in time-series data even in the presence of artifact. It is prevalently used to examine EEG and electrocardiogram (ECG) signals [23]. Permutation entropy [24] measures system complexity by identifying the order relations between time-series values, and obtaining a probability distribution of the ordinal patterns. Sample entropy [24] assists in reflecting and hence diagnosing the unhealthy state of signals. Tsallis entropy [24] is used prevalently for statistical calculations in medicine and physics; hence, it is explored further is this study. Fuzzy entropy [27] computes the level of fuzziness, which is crucial in applied science-related fields; thus, we have also used this parameter. Other parameters include the Kolmogorov–Sinai entropy [28] which controls the maximum amount of information that can be produced by a system, and Kolmogorov complexity [29], a complexed pattern that uses the shortest program as features to describe occurrence. Entropies, such as the modified multiscale entropy [30] which aids in the complexity computation of time-series data, Rényi entropy which is an extension of Shannon’s entropy power, Shannon entropy [32] which typically measures the amount of information required to identify random samples, and wavelet entropy [42] which has the potential to analyze transient features present in nonstationary signals, were also used. Additionally, signal activity [37] which describes the time variation of the signal trace, bispectrum [43], a higher order spectra feature generated by the decomposition of signals, cumulant [44] which is proficient in analyzing imaginary signals, Hjorth parameters [37] which signify statistical properties, recurrence quantitative analysis [45] which computes the number and period of recurrences in a system, largest Lyapunov exponent [46] which detects and distinguishes disorder in a system, and correlation dimension [47] which measures the dimensionality of space occupied by a set of data were also explored. These features have also been used in a study of congestive heart failure detection by Vicnesh et al. [48].
Feature selection and ranking
Highly significant parameters were then chosen using Student’s t test [49]. A t test is a form of statistical test that is performed to compare the mean values between two groups. There are two types of t tests: independent and paired t test. The independent t test is used to compute the difference in mean between two groups that are dependent on each other, while the paired t test is used to compute the difference in mean between two groups that are independent of each other [49]. In this study, the independent t test was used for feature selection. Features of p values greater than 0.05 were discarded, while the remaining features were reduced to a smaller dataset using MFA [50]. MFA is a nonlinear dimensionality reduction technique that is used in areas such as facial recognition. It is a supervised, diverse learning algorithm that works by maintaining the local manifold information. It applies the developments of rank minimisation [51]. MFA is considered to be advantageous as compared to the Linear Discriminant Analysis (LDA) data reduction technique, as it is able to provide the characteristics of intraclass spatial arrangement and interclass disconnectedness. In addition, MFA also has the ability to better characterize the disconnectedness of various classes when compared to the interclass variance in LDA, even without having any prior information regarding data distributions [50]. Thus, MFA is used in this work. The reduced dataset was then subjected to Student’s t test for feature ranking. After feature selection, MFA is employed to condense the large number of features into key components. Hence, this reduction step allows the model to capture all the key information in the data to make accurate predictions. The reduced set is then ranked to obtain the most significant features, necessary for accurate classification. Furthermore, while the reduced dataset still maintains the integrity of the data, it reduces the computational complexity as well. Thus feature selection, MFA, and feature ranking were employed on the entire dataset, before classification. Table 2 presents the best 24 MFA features, in the Appendix section.
Development of a unique autism index
In some of our previous studies, we have successfully developed an index to clearly separate the normal from the abnormal classes [52, 53]. Hence, at this juncture, the significant entropy features were carefully combined to create an index [53] for autism diagnosis, to discern normal from autism class. After various combinations based on the trial-and-error method, the top 3 significant MFA features (MFA 6, MFA 1, MFA 4) were used to develop the index as these features yielded better results and showed the best separation, as compared to the other features that were tested. This is shown in Eq. 1, as established through the development of the index, wherein MFA6, MFA1, and MFA4 refer to the most significant MFA features extracted in this study
Figure 3 shows the boxplot of the developed index, which presents the unique range of values for the diagnosis of normal and autism classes. It is evident from the boxplot that the two classes are well separated, and hence, this attests that the index could be effectively used to discern between the normal and autistic groups. Thus, this not only explains that our features are competent but also that our study is unique and is different from other studies that also explored nonlinear features.
Classification
The optimal feature set was then classified using the SVM classifier with polynomial kernels of order 1, 2, and 3. SVM is highly preferred in classification tasks in machine learning as it produces results of significant accuracy with little computational power. Additionally, SVM is able to better tackle complex classification tasks as compared to other classifiers such as k-nearest neighbor (K-NN) or Naïve Bayes. SVM also has kernel functions which enables it to outperform other classifiers, and it also trains more rapidly. SVM works by designing two hyperplanes that classify all training vectors into 2 groups. The best classification is achieved with the hyperplane that leaves the largest margin between the 2 groups [54]. SVM is prevalently used for the detection of neurological diseases such as epilepsy [55], depression [56], Alzheimer’s disease [57], and schizophrenia [58]. For the above-mentioned reasons, the SVM classifier was considered in this study. K-fold (K = 10) cross-validation [59] was used to evaluate the performance of the classifiers with the aid of the performance metrics: sensitivity, accuracy, specificity, and positive predictive value, wherein 90% of the data were used for training and 10% for testing. Table 1 presents the best classification results achieved with the SVM polynomial 2 classifier.
Results
Comparing the results from Table 1, the SVM polynomial 2 is the best classifier, having the highest accuracy of 98.7%, as compared to the other classifiers used. Since the SVM polynomial 2 classifier was trained and tested with 20 features, it yielded a higher accuracy compared to SVM polynomials 1 and 2. The SVM polynomial 1 classifier achieved the lowest accuracy as it was only trained and tested with eight features. Figure 4 shows the classification accuracies achieved with increasing number of features for the SVM polynomial 2. It can be noted that the highest accuracy was achieved with just 20 MFA features, and it falls further when more than 20 features are taken into consideration. Figure 5a presents scatterplots highlighting the top 3 MFA features. From the scatterplots, it is noticeable that the two classes, normal and autism, are separated, and the features correlate with the classification. Figure 5b presents the scatterplots of the best MFA features. It is apparent that generally, the mean values of MFA features are higher in the normal class, as compared to the autism class. This could be attributed to the higher variance in the normal class. Figure 6a and b illustrates the bispectrum plots for normal and autism signals, respectively. Figures 7a and b illustrates the cumulant plots for normal and autism signals, respectively. From Figs. 5a, 6 and 7, it is obvious that the patterns are very distinguished for the two classes: normal and autism; thus, we have obtained results of high accuracy, due to the extraction of highly discriminant features. Therefore, this affirms the competency and great discriminatory nature of features used for the classification of ASD in our study.
Discussion
CAD systems have been used enthusiastically by other authors to detect ASD. Bhat et al. [60] extracted time and frequency domain features from EEG signals to successfully classify ASD, using data from one healthy subject and one patient. It was established that features extracted from nonlinear analysis can be used as mathematical indicators to detect early stage of ASD. Grobekathofer et al. [61] explored detecting stereotypical motor movements in ASD individuals through the extraction of Recurrence Quantification Analysis (RQA) features, using data from six autistic patients. As the best classifier, random forest yielded an accuracy of more than 86% as compared to SVM, which achieved an accuracy of 86% only.
After pre-processing of EEG signals, Djemal et al. [62] employed the Discrete Wavelet Transform (DWT) to obtain coefficients D1–D4. Entropy and five statistical metrics were then used for feature extraction. The Artificial Neural Network (ANN) was utilized for classification. Data from nine healthy subjects and ten autistic patients were used in this study. DWT together with Shannon entropy yielded the highest accuracy of 99.71% with the ANN classifier.
Acharya et al. [63] discussed research works that employed EEG signals for the diagnosis of autism and the possibility of autism diagnosis through the development of automated tools, using signal processing techniques, in their review paper.
Yousef et al. [64] employed statistical measures with the aid of the Statistical Package for the Social Sciences (SPSS) software to study the relationship between signal patterns and severity of autism. In this study, data were used from 40 healthy subjects and 40 patients. The EEG anomalies correlated with the ASD severity, and the region of brainwave anomalies were observed to be largely correlated to the severity of autism.
Lushchekina et al. [65] obtained EEG recordings using joined ear electrodes with a CONAN 4.5 automated electrophysiological system [66]. The recordings were obtained from 24 normal subjects and 27 early childhood autistic patients. The spectral strength and average coherence were computed in autistic children and compared against normal children. The spectral strength of the theta waves in healthy children was found to be the greatest in the occipital regions, decreasing gradually in the frontal regions.
Elhabashy et al. [67] obtained EEG recordings from children as they rested on a chair with eyes opened. The recordings were obtained from 21 healthy subjects and 21 autistic patients. A 19-electrode cap was fixed on the children according to the 10–20 international standard. Fourier power spectral analysis was conducted on 75 s of artifact-free EEG, which was divided into 4 s epochs. The absolute and relative power were computed for each frequency band per electrode, and coherence indicators were then computed. The absolute power of delta and theta frequency bands were found to be larger in children with ASD, specifically in the frontal region.
Eldridge et al. [68] collected EEG data continuously in a soundproof room as auditory stimuli were presented binaurally to children. The data were obtained from 30 healthy subjects and 19 autistic patients. After pre-processing of signals, the median responses were computed for each participant and the two parameters were compared statistically. The modified multiscale entropy features and variance in time were also extracted from the signals. The SVM, logistic regression, and Naïve Bayes classifiers were employed, with Naïve Bayes yielding the highest accuracy of 79% for classification.
Heunis et al. [69] obtained EEG data from the typical 19 clinical electrode system. The data were obtained from 46 healthy subjects and 16 autistic patients. After pre-processing, the Cross Recurrence Plot toolbox in MATLAB software was then employed to extract ten Recurrence quantification analysis (RQA) features. Tenfold validation and leave-one-out approaches were used for classification. The leave-one-out analysis coupled with SVM delivered the best accuracy of 92.9%.
After the acquisition of signals, Hadoush et al. [70] computed regional averaged MSE values for five cortical regions. In this study, the data were obtained from 18 patients with mid-autism and severe autism each. The MSE curve was plotted to study signal complexity. The SPSS statistical software was employed for quantitative analysis. The Kolmogorov–Smirnov test was used to assess whether the computed multiscale entropy (MSE) values had a normal distribution. The analysis of variance (ANOVA) test was then applied to differentiate the two groups. The averaged MSE values were reported to be larger in mildly autistic children as compared to children with severe autism in the five brain regions assessed.
Buckley et al. [71] extracted features from EEG signals from normal and autistic children in the completely awake, drowsy, and sleep stages. The signals were acquired from 50 healthy subjects (21 with developmental delay without autism and 29 with normal development) and 87 autistic children. The voltage and partial correlations between each electrode pair were recorded to assess functional connectivity. Autistic children were found to exhibit epileptiform activity in the brain signals as compared to the normal children.
Pham et al. [72] obtained EEG signals from 37 healthy subjects and 40 autistic patients and employed higher order spectra (HOS) bispectrum to convert the acquired signals to images and extracted nonlinear features from the images. The locality sensitivity discriminant analysis (LSDA) was then used to condense the features. Highly discriminatory features were then chosen using the t test, and fed to various classifiers subsequently. The classifiers were evaluated using tenfold cross-validation. The PNN classifier achieved the highest accuracy of 98.70%, outperforming the other classifiers.
Cheong et al. [73] obtained EEG signals from 30 autistic children and pre-processed the signals using the discrete wavelet transform and extracted nonlinear features. These were then classified with the multilayer perceptron model, yielding a classification accuracy of 92.3%.
Grossi et al. [74] embarked on a pilot study and explored a newer IFAST algorithm together with multiscale entropy features and ranked organizing maps for the classification of autism. In this study, data were obtained from 10 healthy subjects and 15 autistic patients. An accuracy of 100% was achieved for the training and testing validation method, while an accuracy between 84% and 92.8% was achieved for the leave-one-out validation method.
Sinha et al. [75] obtained EEG signals from 20 healthy subjects and 10 autistic patients and pre-processed the acquired signals using the discrete wavelet transform and extracted nonlinear features thereafter. These features were fed to the K-NN classifier, yielding a classification accuracy of 92.8%.
Haputhanthri [76] obtained EEG signals from 10 healthy subjects and 10 autistic patients and extracted statistical features after pre-processing the signals using the discrete wavelet transform (DWT) technique. Feature selection was then done based on correlation, after which the features were input to the random forest classifier, achieving an accuracy of 93%.
Abdolzadegan et al. [71] obtained EEG signals from 11 healthy subjects and 34 autistic patients and extracted a combination of linear and nonlinear parameters from the signals. The features were fed to some classifiers wherein the SVM classifier achieved the highest classification accuracy of 90.57%.
Arunkumar et al. [77] obtained EEG signals from 4 healthy subjects and 6 autistic patients and explored spectral analysis of the signals using the short-time Fourier transform. Through this study, it was reported that EEG signals could be used to successfully classify autism versus normal.
Subudhi et al. [78] acquired EEG signals from 41 autistic patients and 32 healthy subjects and extracted nonlinear features from the signals. The significant features were classified thereafter using the support vector machine classifier, yielding an accuracy of 90.41%.
Tawhid et al. [79] acquired EEG signals from 12 autistic patients and 4 healthy subjects and transformed the 1-dimensional signals to 2-dimensional spectrogram images using Fourier transform. Significant features are selected using principal component analysis thereafter, and fed to the support vector machine classifier for classification, yielding an accuracy of 95.25%.
A slightly different study was conducted by Bosl et al. [80], for predicting autism risk. After acquiring the EEG data, the signal or time-series was decomposed into different frequency bands using the Daubechies (DB4) wavelet transform, and nine nonlinear features were obtained from the frequency bands. Discriminatory features were chosen using the recursive feature elimination algorithm, and classification was performed thereafter using SVM with radial basis functions. With the proposed method, high specificity, sensitivity, and positive predictive values of > 95% were yielded for some ages.
It is notable from the discussion above that most of the studies employed nonlinear feature extraction to study the physiological conditions of the EEG signals and cross-validation techniques to evaluate proposed methods. Since EEG signals are time-varying and disordered naturally; nonlinear features were also exploited for the classification of autism in this study.
EEG signals are generally nonlinear, and spiking patterns of the brain signals provide useful information about brain conditions. Hence, EEG signals are studied fervently using nonlinear feature extraction methods. As these biosignals are also multiscaled, the signals display different behaviors, depending on the scale at which they are being examined, potentially providing insights into different types of diseases. Multiple recent studies have analyzed EEG signals using multiscale entropy. Our proposed CAD system employs feature extraction methods for the classification of brain signals into ASD and normal EEG signals. The SVM polynomial 2 performs the best with 20 features, compared to the other two classifiers explored, yielding the highest accuracy of 98.7%. Comparing the studies as discussed above, Bhat et al. [60] have used nonlinear features for classification, but the data size is insignificant for comparison. Eldridge et al. [68] extracted the modified multiscale entropy feature from EEG signals, but the classification accuracy achieved was lower than in our study. Hadoush et al. [70] studied multiscale entropy features, but only two features were extracted, without any classification. Furthermore, classification was not done. Heunis et al. [69] performed RQA nonlinear analysis of signals. However, lesser features were used, and the classification accuracy achieved was lower than in our study. Grobekathofer et al. [61] also explored RQA feature extraction methods; however, the data size was small (n = 6). Furthermore, the study is not as promising, since normal subjects were not included.
Lushchekina et al. [65], Elhabashy et al. [67], and Yousef et al. [64] employed statistical analysis methods. However, the spectral and absolute power in specific brain regions of autistic children as well as correlations of autism severity with EEG abnormality were reported, but classification was not done. Djemal et al. [62] extracted entropy features and achieved a slightly higher accuracy than our study, but the data size is inconsiderable by comparison. Buckley et al. [71] reported the relationship between epileptic activity in EEG signals of autistic children and that of normal children, but a classification was not done. Arunkumar et al. [77] did not do a classification study and reported on qualitative results. Grossi et al. [74] and Haputhanthri. [76] had achieved lower accuracies than our study, for their classification methods. Although Abdolzadegan et al. [71] had explored nonlinear features similar to our study, linear features were also extracted, wherein a lower accuracy was obtained as compared to our study. Cheong et al. [73] and Sinha et al. [53] had also extracted nonlinear features similar to our study, but obtained lower accuracies than our study. Although Subudhi et al. [78] and Tawhid et al. [79] had obtained higher accuracies than our study, the authors had used lesser data as compared to our study. Pham et al. [72] did a similar classification to that of our study and achieved the same accuracy as ours (98.7%). However, Pham et al. [72] extracted the HOS bispectrum and nonlinear texture features and employed LSDA, while in our study, we extracted different nonlinear features and had employed MFA. Furthermore, we uniquely developed an ASD index. Hence, it is evident that our proposed method is unique, rapid, and the most expedient, to be considered as a potential diagnostic tool for the classification of ASD by healthcare professionals. The following are the advantages and disadvantages of this work:
Advantages
-
i.
The proposed system is robust, as it has been validated by tenfold cross-validation.
-
ii.
The proposed technique is a novel classification technique for autism. The high accuracy obtained is due to the highly discriminative feature set used.
-
iii.
Rapid and accurate diagnosis of autism is possible with the developed classification system.
-
iv.
We are one of the first groups to have developed a novel ASD index in this study, wherein ASD can be effectively diagnosed using a single number.
-
v.
Unique bispectrum and cumulant plots have been presented for normal and ASD classes.
Disadvantages
-
i.
The feature extraction process is done manually and the optimal features are selected by hand, making it cumbersome as a whole.
-
ii.
The data used are insufficient to train a deep learning model for classification.
Conclusion
In this study, a unique classification system was developed to categorize autism and normal EEG signals. Eighteen nonlinear features were extracted from the acquired brain signals. Highly significant features were then selected based on the t test. The significant feature set was then reduced using MFA before being ranked based on the t values from the t test. The reduced, significant feature set was then input to SVM polynomials 1, 2, and 3, while the three most significant features were used to develop an index for autism diagnosis. Tenfold was used to validate our classification system. A high classification accuracy of 98.70% was achieved with 20 features using the SVM polynomial 2 classifier. This attests that the proposed technique is effective in the classification of normal versus ASD children. Although the proposed method is promising, manually extracting features can be cumbersome. To mitigate, we intend to gather more data and employ deep learning algorithms for ASD diagnosis. We also propose to include the cloud-based detection model as part of our future work.
Future work
To enhance the efficiency of our system, we recommend adding a cloud-based detection element. Figure 8 illustrates how the proposed cloud-based detection system works. The EEG signals extracted from patients would be stored in the hospital’s server and sent to cloud, where our developed CAD system would be located. Upon analysis, the classification result would be sent back to the server to aid the clinician in decision-making. The clinician would then send the results back to the server, which would communicate directly with the patient smartphone. With the application of this system, the clinician's task would be simplified. We also intend to test the model with additional EEG signals, and upload to cloud for faster diagnosis as part of our future work. With more data available, blindfold validation would become useful to incorporate into the system. With the data used to train the model each time, it will become more robust. With more data available, deep learning techniques could also be explored. The advantage of placing the proposed system in cloud is that it could also be used to diagnose other neurological diseases that affect children, such as attention-deficit hyperactivity disorder and epilepsy.
References
Spitzer RL, Skodol AE, Gibbon M, Williams JBW (1985) Diagnostic and statistical manual of mental disorders, 3rd edition, vol 15, no 3, pp 703–704
Kim DG, Park HR, Lee JM, Moon HE, Lee DS, Kim BN, Kim J, Paek SH (2016) A short review on the current understanding of autism spectrum disorders. Exp Neurobiol 25(1):1–13
Miles JH (2011) Autism spectrum disorders-A genetics review. Genet Med 13(4):278–294
Szatmari P, Jones MB, Zwaigenbaum L, MacLean JE (1998) Genetics of autism: overview and new directions. J Autism Dev Disord 28(5):351–368
London E, Etzel RA (2000) The environment as an etiologic factor in autism: a new direction for research. Environ Health Perspect 108(SUPPL. 3):401–404
Barnes JC, TenEyck M (2017) Prenatal and Perinatal Risk Factors for Delinquency. Encycl Juv Delinq Justice 161:1–6
Ashwood P, Van De Water J (2004) Is autism an autoimmune disease? Autoimmun Rev 3(7–8):557–562
Davidson PW, Myers GJ, Weiss B (2004) Mercury exposure and child development outcomes. Pediatrics 113(4 Suppl):1023–1029
Sharif H, Khan RA (2019) A novel framework for automatic detection of Autism: a study on Corpus Callosum and Intracranial Brain Volume
Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D (2014) Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83(5):1131–1143
Onaolapo Y, Onaolapo O (2017) Global data on autism spectrum disorders prevalence: a review of facts, fallacies and limitations. Univers J Clin Med 5(2):14–23
Kabot S, Masi W, Segal M (2003) Advances in the diagnosis and treatment of autism spectrum disorders. Prof Psychol Res Pract 34(1):26–33
Oh SL, Hagiwara Y, Raghavendra U, Yuvaraj R, Arunkumar N, Murugappan M, Acharya UR (2018) A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput Appl 32:10927–10933
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H, Subha DP (2018) Automated EEG-based screening of depression using deep convolutional neural network. Comput Methods Programs Biomed 161:103–113
Acharya UR, Hagiwara Y, Deshpande S, Suren S, Koh JEW, Oh SL, Arunkumar N, Ciaccio EJ, Lim CM (2019) Characterization of focal EEG signals: a review. Fut Gen Comput Syst 91:290–299
Bhat S, Acharya UR, Adeli H, Bairy GM, Adeli A (2014) Autism: cause factors, early diagnosis and therapies. Rev Neurosci 25(6):56
Hadoush H, Alafeef M, Abdulhay E (2019) Brain complexity in children with mild and severe autism spectrum disorders: analysis of multiscale entropy in EEG. Brain Topogr 32(5):914–921
Hadoush H, Alafeef M, Abdulhay E (2019) Automated identification for autism severity level: EEG analysis using empirical mode decomposition and second order difference plot. Behav Brain Res 362:240–248
Jahmunah V, Lih S, Rajinikanth V, Ciaccio EJ, Hao K (2019) Automated detection of schizophrenia using nonlinear signal processing methods. Artif Intell Med 100:101698
Loo SK, Makeig S (2012) Clinical utility of EEG in attention-deficit/hyperactivity disorder: a research update. Neurotherapeutics 9(3):569–587
Houmani N, Vialatte F, Jutgla EG, Dreyfus G, Michel VHN, Mariani J, Kinugwa K (2018) Diagnosis of Alzheimer’s disease with Electroencephalography in a differential framework. PLoS ONE 13(3):e0193607–e0193607
Lo MT, Tsai PH, Lin PF, Lin C, Hsin YL (2009) The nonlinear and nonstationary properties in EEG signals: Probing the complex fluctuations by Hilbert-huang transform. Adv Adapt Data Anal 1(3):461–482
Pincus SM (1991) Approximate entropy as a measure of system complexity (stadstc/stohastdc processes/chaos/dimension). Proc Nati Acad Sci USA 88(March):2297–2301
Dostál O, Vysata O, Pazdera L, Prochazka A, Kpoal J, Kuchynka J, Valis M (2018) Permutation entropy and signal energy increase the accuracy of neuropathic change detection in needle EMG. Comput Intell Neurosci 2018:1–5
Richman JS, Moorman JR (2017) Physiological time-series analysis using approximate entropy and sample entropy. Am J Physiol Circ Physiol 278(6):H2039–H2049
Anastasiadis A (2012) Special issue: Tsallis entropy. Entropy 14(2):174–176
Ramer A (1990) Concepts of fuzzy information measures on continuous domains. Int J Gen Syst 17(2–3):241–248
Pham TD (2016) The Kolmogorov-Sinai entropy in the setting of fuzzy sets for image texture analysis and classification. Pattern Recognit 53:229–237
Kolmogorov AN (1968) Three approaches to the quantitative definition of information. Int J Comput Math 2(1–4):157–168
De Wu S, Wu CW, Lee KY, Lin SG (2013) Modified multiscale entropy for short-term time series analysis. Phys A Stat Mech Appl 392(23):5865–5873
Savare G, Toscani G (2014) The concavity of rényi entropy power. IEEE Trans Inf Theory 60(5):2687–2693
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27(1924):623–656
Rosso OA, Blanco S, Yordanova J, Kolev V, Figliola A, Schuar M (2001) Wavelet entropy: a new tool for analysis of short duration brain electrical signals. Neurosci Method 105:65–75
Sweller J, Van Merrienboer JJG, Paas FGWC (1998) Cognitive architecture and instructional design. Educ Psychol Rev 10(3):251–296
Collis WB, White PR, Hammond JK (1998) A mathematical theory of communication. Mech Syst Signal Process 12(3):375–394
Brillinger DR (1965) An introduction to polyspectra. Ann Math Stat 36(5):1351–1374
Hjorth B (1970) EEG analysis based on time domain properties. Electroencephalogr Clin Neurophysiol 29:306–310
Curtin PCP, Curtin A, Austin C, Gennings C, Tammimies K, Bolte S, Arora M (2017) Recurrence quantification analysis to characterize cyclical components of environmental elemental exposures during fetal and postnatal development. PLoS ONE 12(11):0187049
Pavlov AN, Pavlova ON, Kurths J (2017) Determining the largest Lyapunov exponent of chaotic dynamics from sequences of interspike intervals contaminated by noise. Eur Phys J B 90(4):70439
Gong Z (2015) A super-high-efficiency algorithm for the calculation of the correlation integral. J Data Anal Inf Process 03(04):128–135
Rényi A (1961) On measures of entropy and information. Fourth Berkeley Symp Math Stat Probab 1:547–561
Rosso OA, Yordanova SBJ, Kolev V, Figliola A, Schurmann M, Basar E (2001) Wavelet entropy: a new tool for analysis of short duration brain electrical signals. J Neurosci Methods 105(1):65–75
Nikias CL, Raghuveer MR (1987) Bispectrum estimation: A digital signal processing framework. Proc IEEE 75(7):869–891
Acharya UR, Sree SV, Suri JS (2011) Automatic detection of epileptic eeg signals using higher order cumulant features. Int J Neural Syst 21(5):403–414
Marwan N, Webber CL (2015) Mathematical and computational foundations of recurrence quantifications. https://doi.org/10.1007/978-3-319-07155-8_1
Liu H-F, Yang Y-Z, Dai Z-H, Yu Z-H (2003) The largest Lyapunov exponent of chaotic dynamical system in scale space and its application. Chaos An Interdiscip J Nonlinear Sci 13(3):839–844
Peter Grassberger IP (1983) Measuring the strangeness of strange attractors. Physica D 9(1–2):189–208
Jahmunah V, Oh SL, Koh JEW, Ciaccio EJ, Chua KC, Tan RS (2019) Computer-aided diagnosis of congestive heart failure using ECG signals: a review. Phys Med 62:95–104
Kim TK (2015) T test as a parametric statistic. Korean J Anesthesiol 68(6):540–546
Chang Y (2014) Graph embedding and extensions: a general framework for dimensionality reduction. IEEE Trans Pattern Anal Mach Intell 29(1):40–51
Wang Z, Sun X, Sun L, Huang Y (2013) Semisupervised kernel marginal Fisher analysis for face recognition. Sci World J 2013:1–13
Acharya UR, Faust O, Subbhuraam VS, Ghista DN, Dua S, Joseph P, Thajudin A, Janarthanan N, Tamura T (2013) An integrated diabetic index using heart rate variability signal features for diagnosis of diabetes. Comput Methods Biomech Biomed Engin 16(2):222–234
Raghavendra U, Pham TH, Gudigar A, Vidhya V, Rao BN, Sabut S, Koh JEW, Ciaccio EJ, Acharya UR (2021) Novel and accurate non-linear index for the automated detection of haemorrhagic brain stroke using CT images. Complex Intell Syst 7:929–940
S. V. Machines, “C o m p a r i son o f C l a ss i fi e r s,” .
Tang Y, Durand DM (2012) A tunable support vector machine assembly classifier for epileptic seizure detection. Expert Syst Appl 39:3925–3938
Acharya UR, Sudarshan VK, Adeli H, Santhosh J, Koh JEW, Putahnkatti SD, Adeli A (2015) A novel depression diagnosis index using nonlinear features in EEG signals. Eur Neurol 74(1–2):79–83
Rabeh AB, Benzarti F, Amiri H (2016) Diagnosis of Alzheimer diseases in early step using SVM (Support Vector Machine). In: 2016 13th International conference on computer graphics, imaging and visualization (CGiV), 2016, pp 364–367
Jahmunah V, Oh SL, Rajinikanth V, Ciaccio EJ, Cheong KH, Arunkumar N, Acharya UR (2019) Automated detection of schizophrenia using nonlinear signal processing methods. Artif Intell Med 100:101698
Duda R, Hart R, Stork D (2012) Pattern classification
Bhat S, Acharya UR, Adeli H, Bairy GM, Adeli A (2014) Automated diagnosis of autism: in search of a mathematical marker. Rev Neurosci 25(6):36
Großekathöfer U, Manyakov N, Mihajlovic V, Pandina GJ, Skalkin A, Ness S, Bangerter A, Goodwin MS (2017) Automated detection of stereotypical motor movements in autism spectrum disorder using recurrence quantification analysis. Front Neuroinform 11:6
Djemal R, AlSharabi K, Ibrahim S, Alsuwailem A (2017) EEG-based computer aided diagnosis of autism spectrum disorder using wavelet, entropy, and ANN. Biomed Res Int 2017:1–9
Bhat S, Acharya UR, Adeli H, Bairy GM, Adeli A (2014) Autism: Cause factors, early diagnosis and therapies. Rev Neurosci 25(6):841–850
Yousef A, Youssef U, El-Shabrawy A, Abdel Fattah NA, Khedr H, Khedr H (2017) EEG abnormalities and severity of symptoms in non-epileptic autistic children. Egypt J Psychiatry 38(2):59
Lushchekina EA, Podreznaya ED, Lushchekin VS, Novototskii-Vlasov VY, Strelets VB (2014) Comparative studies of EEG theta and gamma rhythms in normal children and children with early childhood Autism. Neurosci Behav Physiol 44(8):902–908
Kulaichev AP (2002) Computerized Electrophysiology. Moscow State University Press, Moscow
Elhabashy H, Raafat O, Afifi L, Raafat H, Abdullah K (2015) Quantitative EEG in autistic children. Egypt J Neurol Psychiatry Neurosurg 52(3):176–182
Eldridge J, Lane AE, Belkin M, Dennis S (2014) Robust features for the automatic identification of autism spectrum disorder in children. J Neurodev Disord 6(1):1–12
Heunis T, Aldrich C, Peters JM, Jeste SS, Sahin M, Scheffer C, de Vries PJ (2018) Recurrence quantification analysis of resting state EEG signals in autism spectrum disorder - a systematic methodological exploration of technical and demographic confounders in the search for biomarkers. BMC Med 16(1):1–17
H. Hadoush, M. Alafeef, E. Abdulhay, Brain complexity in children with mild and severe autism spectrum disorders: analysis of multiscale entropy in EEG. Brain Topogr., no. 0123456789, 2019.
Abdolzadegan D, Moattar MH, Ghoshuni M (2020) A robust method for early diagnosis of autism spectrum disorder from EEG signals based on feature selection and DBSCAN method. Biocybern Biomed Eng 40(1):482–493
Pham T-H, Vicnesh J, Wei JKE, Oh SL, Arunkumar N, Abdulhay EW, Ciaccio EJ, Acharya UR (2020) Autism spectrum disorder diagnostic system using {HOS} bispectrum with {EEG} signals. Int J Environ Res Public Health 17:971. https://doi.org/10.3390/ijerph17030971
Cheong LC, Sudirman R, Hussin SS (2015) Feature extraction of EEG signal using wavelet transform for autism classification. ARPN J Eng Appl Sci 10(19):8533–8540
Grossi E, Olivieri C, Buscema M (2017) Diagnosis of autism through EEG processed by advanced computational algorithms: A pilot study. Comput Methods Programs Biomed 142:73–79
Sinha T, Munot MV, Sreemathy R (2019) An efficient approach for detection of autism spectrum disorder using electroencephalography signal. IETE J Res 2063:1–9
D. Haputhanthri, G. Brihadiswaran, S. Gunathilaka, D. Meedeniya, Y. Jayawardana, S. Jayarathna, M. Jaime, “An EEG based Channel Optimized Classification Approach for Autism Spectrum Disorder,” MERCon 2019 - Proceedings, 5th Int. Multidiscip. Moratuwa Eng. Res. Conf., pp. 123–128, 2019.
Arunkumar B, Vijay SAA, Kumar KS (2020) Spectral analysis based differentiationfor EEG signals of children with autism. J Crit Rev 7(4):882–887
Subudhi AK, Mohanty M, Sahoo SK, Mohanty SK, Mohanty B (2020) Automated delimitation and classification of autistic disorder using EEG signal. IETE J Res 2020:1–9
Tawhid MNA, Siuly S, Wang H (2020) Diagnosis of autism spectrum disorder from EEG using a time-frequency spectrogram image-based approach. Electron Lett 56(25):1372–1375
Bosl WJ, Tager-Flusberg H, Nelson CA (2018) EEG analytics for early detection of autism spectrum disorder: a data-driven approach. Sci Rep 8(1):1–20
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oh, S.L., Jahmunah, V., Arunkumar, N. et al. A novel automated autism spectrum disorder detection system. Complex Intell. Syst. 7, 2399–2413 (2021). https://doi.org/10.1007/s40747-021-00408-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00408-8