Abstract
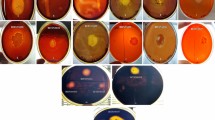
Although several experimental works have been focused on landfill waste/wastewater treatment, no research has been carried out to evaluate the major enzyme activities expressed by indigenous microbiota. In this work, a total of 30 bacterial strains were isolated from a recent operating landfill stabilization pond and were identified by 16S rRNA gene sequencing. The majority of the isolates were phylogenetically related to different species of the genera Paracoccus, Pseudomonas and Aquamicrobium, whereas two bacterial isolates were distantly related to the species Gemmobacter aquatilis and Paenirhodobacter enshiensis (95.1 and 96.3 % similarity, respectively). All the bacterial isolates obtained from the stabilization pond were further examined for their endo- and exocellular β-glucosidase, lipase and protease activities. Protease activities were either undetected or extremely low (below 10 U g−1 protein), while lipase activities were exhibited by only certain bacterial strains. However, almost all bacterial isolates exhibited β-glucosidase activities (half of the isolates exerting activities above 100 U g−1 protein), indicating their β-glycosidic nature. Indeed, the presence of an important cellulose fraction in the effluent may explain the high β-glucosidase activities expressed by the majority of the bacterial strains.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Andreoni V, Cavalca L, Rao MA, Nocerino G, Bernasconi S, Dell’Amico E, Colombo M, Gianfreda L (2004) Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 57:401–412
Arenghi FLG, Berlanda D, Galli E, Sello G, Barbieri P (2001) Organization and regulation of meta cleavage pathway genes for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl Environ Microbiol 67:3304–3308
Bambauer A, Rainey FA, Stackebrandt E, Winter J (1998) Characterization of Aquamicrobium defluvii gen. nov. sp. nov., a thiophene-2-carboxylate-metabolizing bacterium from activated sludge. Arch Microbiol 169:293–302
Barlaz MA, Schaefer DM, Ham RK (1989) Bacterial population development and chemical characteristics of refuse decomposition in a simulated sanitary landfill. Appl Environ Microbiol 55:55–65
Benitez E, Sainz H, Nogales R (2005) Hydrolytic enzyme activities of extracted humic substances during the vermicomposting of a lignocellulosic olive waste. Bioresour Technol 96:785–790
Bhatia Y, Mishra S, Bisaria VS (2002) Microbial β-glucosidases: cloning, properties, and applications. Crit Rev Biotechnol 22:375–407
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burrell PC, O’Sullivan C, Song H, Clarke WP, Blackall LL (2004) Identification, detection, and spatial resolution of Clostridium populations responsible for cellulose degradation in a methanogenic landfill leachate bioreactor. Appl Environ Microbiol 70:2414–2419
Busquets A, Peña A, Gomila M, Bosch R, Nogales B, García-Valdé E, Lalucat J, Bennasar A (2012) Genome sequence of Pseudomonas stutzeri strain JM300 (DSM 10701), a soil isolate and model organism for natural transformation. J Bacteriol 194:5477–5478
Calli B, Tas N, Mertoglu B, Inanc B, Ozturk I (2003) Molecular analysis of microbial communities in nitrification and denitrification reactors treating high ammonia leachate. J Environ Sci Health A Tox Hazard Subst Environ Eng 38:1997–2007
Chang Y-C, Takada K, Choi D, Toyama T, Sawada K, Kikuchi S (2013) Isolation of biphenyl and polychlorinated biphenyl-degrading bacteria and their degradation pathway. Appl Biochem Biotechnol 170:381–398
Chien PCT, Yoo H-S, Dykes GA, Lee SM (2015) Isolation and characterization of cellulose degrading ability in Paenibacillus isolates from landfill leachate. Malays J Microbiol 11:185–194
Chua P, Yoo H-S, Gan HM, Lee S-M (2014) Draft genome sequences of two cellulolytic Paenibacillus sp. strains, MAEPY1 and MAEPY2, from Malaysian landfill leachate. Genome Announc 2:e00065–14
Clesceri LS, Greenberg AE, Eaton AD (1998) Standard methods for the examination of water and wastewater. American Public Health Association (APHA), Washington DC
Etchebehere C, Errazquin I, Barrandeguy E, Dabert P, Moletta R, Muxi L (2001) Evaluation of the denitrifying microbiota of anoxic reactors. FEMS Microbiol Ecol 35:259–265
Gessesse A, Dueholm T, Petersen SB, Nielsen PH (2003) Lipase and protease extraction from activated sludge. Water Res 37:3652–3657
Guo L, Ji M, Dong H, Wei Y (2009) Screening and degradation performances of dominant strains in high-salinity landfill leachate. Appl Microbiol Biotechnol 84:357–364
Hata J, Miyata N, Kim E-S, Takamizawa K, Iwahori K (2004) Anaerobic degradation of cis-1,2-dichloroethylene and vinyl chloride by Clostridium sp. strain DC1 isolated from landfill leachate sediment. J Biosci Bioeng 97:196–201
Heyrman J, Logan NA, Rodríguez-Díaz M, Scheldeman P, Lebbe L, Swings J, Heyndrickx M, De Vos P (2005) Study of mural painting isolates, leading to the transfer of ‘Bacillus maroccanus’ and ‘Bacillus carotarum’ to Bacillus simplex, emended description of Bacillus simplex, re-examination of the strains previously attributed to ‘Bacillus macroides’ and description of Bacillus muralis sp. nov. Int J Syst Evol Microbiol 55:119–131
Hoornweg D, Bhada-Tata P, Kennedy C (2013) Environment: waste production must peak this century. Nature 502:615–617
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huang L-N, Zhou H, Zhu S, Qu L-H (2004) Phylogenetic diversity of bacteria in the leachate of a full-scale recirculating landfill. FEMS Microbiol Ecol 50:175–183
Huber B, Drewes JE, Lin KC, König R, Müller E (2014) Revealing biogenic sulfuric acid corrosion in sludge digesters: detection of sulfur-oxidizing bacteria within full-scale digesters. Water Sci Technol 70:1405–1411
Huset CA, Barlaz MA, Barofsky DF, Field JA (2011) Quantitative determination of fluorochemicals in municipal landfill leachates. Chemosphere 82:1380–1386
Islas-Espinoza M, Reid BJ, Wexler M, Bond PL (2012) Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure. Microb Ecol 64:140–151
Jaeger K-E, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic, New York, pp 21–132
Karak T, Bhagat RM, Bhattacharyya P (2012) Municipal solid waste generation, composition, and management: the world scenario. Crit Rev Environ Sci Technol 42:1509–1630
Kobayashi T, Koide O, Mori K, Shimamura S, Matsuura T, Miura T, Takaki Y, Morono Y, Nunoura T, Imachi H, Inagaki F, Takai K, Horikoshi K (2008) Phylogenetic and enzymatic diversity of deep subseafloor aerobic microorganisms in organics- and methane-rich sediments off Shimokita Peninsula. Extremophiles 12:519–527
Kong J-Y, Su Y, Zhang Q-Q, Bai Y, Xia F-F, Fang C-R, He R (2013) Vertical profiles of community and activity of methanotrophs in landfill cover soils of different age. J Appl Microbiol 115:756–765
Li T, Mazéas L, Sghir A, Leblon G, Bouchez T (2009) Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11:889–904
Li L, Han Y, Yan X, Liu J (2013) H2S removal and bacterial structure along a full-scale biofilter bed packed with polyurethane foam in a landfill site. Bioresour Technol 147:52–58
Lipski A, Kämpfer P (2012) Aquamicrobium ahrensii sp. nov. and Aquamicrobium segne sp. nov., isolated from experimental biofilters. Int J Syst Evol Microbiol 62:2511–2516
Lipski A, Reichert K, Reuter B, Spröer C, Altendorf K (1998) Identification of bacterial isolates from biofilters as Paracoccus alkenifer sp. nov. and Paracoccus solventivorans with emended description of Paracoccus solventivorans. Int J Syst Bacteriol 48:529–536
Liu J, Wu W, Chen C, Sun F, Chen Y (2011) Prokaryotic diversity, composition structure, and phylogenetic analysis of microbial communities in leachate sediment ecosystems. Appl Microbiol Biotechnol 91:1659–1675
Liu M, Zhang Y, Ding R, Gao Y, Yang M (2013) Response of activated sludge to the treatment of oxytetracycline production waste stream. Appl Microbiol Biotechnol 97:8805–8812
Mormile MR, Gurijala KR, Robinson JA, McInerney MJ, Suflita JM (1996) The importance of hydrogen in landfill fermentations. Appl Environ Microbiol 62:1583–1588
Ohnishi A, Nagano A, Fujimoto N, Suzuki M (2011) Phylogenetic and physiological characterization of mesophilic and thermophilic bacteria from a sewage sludge composting process in Sapporo, Japan. World J Microbiol Biotechnol 27:333–340
Peeters K, Hodgson DA, Convey P, Willems A (2011) Culturable diversity of heterotrophic bacteria in Forlidas pond (Pensacola mountains) and Lundström Lake (Shackleton range), Antarctica. Microb Ecol 62:399–413
Qiu Z, Liu Y, Wang Q, Tang J, Tong F, Zhou L (2011) Screening functional strains and constructing complex microbial system for accelerating stabilization of landfill. J Southwest Jiaotong Univ 46:706–711
Qu G, de Varennes A (2010) Use of hydrophilic polymers from diapers to aid the establishment of Spergularia purpurea in a mine soil. J Hazard Mater 178:956–962
Rainey FA, Klatte S, Kroppenstedt RM, Stackebrandt E (1995) Dietzia, a new genus including Dietzia maris comb. nov., formerly Rhodococcus maris. Int J Syst Bacteriol 45:32–36
Rao MB, Tanksale AM, Ghatge MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Rashamuse K, Magomani V, Ronneburg T, Brady D (2009) A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous β-lactamase activity on nitrocefin. Appl Microbiol Biotechnol 83:491–500
Renou S, Givaudan JG, Poulain S, Dirassouyan F, Moulin P (2008) Landfill leachate treatment: review and opportunity. J Hazard Mater 150:468–493
Rothe B, Fischer A, Hirsch P, Sittig M, Stackebrandt E (1987) The phylogenetic position of the budding bacteria Blastobacter aggregatus and Gemmobacter aquatilis gen., nov. sp. nov. Arch Microbiol 147:92–99
Rüling WF, Van Breukelen BM, Braster M, Van Verseveld HW (2000) Linking microbial community structure to pollution: biolog-substrate utilization in and near a landfill leachate plume. Water Sci Technol 41:47–53
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sawamura H, Yamada M, Endo K, Soda S, Ishigaki T, Ike M (2010) Characterization of microorganisms at different landfill depths using carbon-utilization patterns and 16S rRNA gene based T-RFLP. J Biosci Bioeng 109:130–137
Semrau JD (2011) Current knowledge of microbial community structures in landfills and its cover soils. Appl Microbiol Biotechnol 89:961–969
Sharma D, Sharma B, Shukla AK (2011) Biotechnological approach of microbial lipase: a review. Biotechnology 10:23–40
Shen D-S, He R, Ren G-P, Traore I, Feng X-S (2002) Effect of leachate recycling and inoculation on the biochemical characteristics of municipal refuse in landfill bioreactors. J Environ Sci 14:406–412
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7:539
Siller H, Rainey FA, Stackebrandt E, Winter J (1996) Isolation and characterization of a new gram-negative, acetone-degrading, nitrate-reducing bacterium from soil, Paracoccus solventivorans sp. nov. Int J Syst Bacteriol 46:1125–1130
Singh V, Mittal AK (2013) Performance aspects of Paracoccus pantotrophus treating urban solid waste leachate. Desalin Water Treat 51(10–12):2474–2479
Song L, Wang Y, Tang W, Lei Y (2015) Bacterial community diversity in municipal waste landfill sites. Appl Microbiol Biotechnol 99:7745–7756
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tanaka Y, Hanada S, Manome A, Tsuchida T, Kurane R, Nakamura K, Kamagata Y (2004) Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth. Int J Syst Evol Microbiol 54:955–959
Tiwari R, Pranaw K, Singh S, Nain PK, Shukla P, Nain L (2015) Two-step statistical optimization for cold active β-glucosidase production from Pseudomonas lutea BG8 and its application for improving saccharification of paddy straw. Biotechnol Appl Biochem. doi:10.1002/bab.1415
Wang D, Liu H, Zheng S, Wang G (2014) Paenirhodobacter enshiensis gen. nov., sp. nov., a non-photosynthetic bacterium isolated from soil, and emended descriptions of the genera Rhodobacter and Haematobacter. Int J Syst Evol Microbiol 64:551–558
Wei Y, Zhou H, Zhang J, Zhang L, Geng A, Liu F, Zhao G, Wang S, Zhou Z, Yan X (2015) Insight into dominant cellulolytic bacteria from two biogas digesters and their glycoside hydrolase genes. PLoS ONE 10:e0129921
Westlake K (1995) Landfill waste pollution and control. Albion Publishing, Chichester, UK
Woo S-G, Srinivasan S, Yang J, Jung Y-A, Kim MK, Lee M (2012) Nocardioides daejeonensis sp. nov., a denitrifying bacterium isolated from sludge in a sewage disposal plant. Int J Syst Evol Microbiol 62:1199–1203
Yoon J-H, Lee C-H, Oh T-K (2005) Nocardioides dubius sp. nov., isolated from an alkaline soil. Int J Syst Evol Microbiol 55:2209–2212
Yu G, He P, Shao L, Lee D (2008) Extracellular enzymes in sludge flocs collected at 14 full-scale wastewater treatment plants. J Chem Technol Biotechnol 83:1717–1725
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ntougias, S. Phylogenetic Identification and Enzyme Activities of Indigenous Bacteria from a Landfill Stabilization Pond. Environ. Process. 3, 341–352 (2016). https://doi.org/10.1007/s40710-016-0150-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40710-016-0150-6