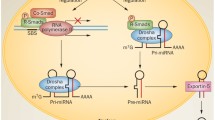
Abstract
IgA nephropathy (IgAN) is one of the most common glomerulonephritides. The disease is characterized by haematuria, proteinuria, deposition of galactose-deficient IgA1 in the glomerular mesangium and mesangial hypercellularity, further leading to extracellular matrix expansion. Kidney biopsy is the gold standard for IgAN diagnosis. Due to the invasiveness of renal biopsy, there is an unmet need for noninvasive biomarkers to diagnose and estimate the severity of IgAN. Understanding the role of RNA molecules as genetic markers to target diseases may allow developing therapeutic and diagnostic markers. In this review we have focused on intrarenal, extrarenal and extracellular noncoding RNAs involved in the progression of IgAN. This narrative review summarizes the pathogenesis of IgAN along with the correlation of noncoding RNA molecules such as microRNAs, small interfering RNAs, circular RNAs and long non-coding RNAs that play an important role in regulating gene expression, and that represent another type of regulation affecting the expression of specific glycosyltranferases, a key element contributing to the development of IgAN.
Graphical abstract




Similar content being viewed by others
References
Schena FP, Nistor I (2018) Epidemiology of IgA nephropathy: a global perspective. Semin Nephrol 38(5):435–442. https://doi.org/10.1016/j.semnephrol.2018.05.013
McGrogan A, Franssen CF, de Vries CS (2011) The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol Dial Transpl 26(2):414–430. https://doi.org/10.1093/ndt/gfq665
Berthoux F, Mohey H, Laurent B, Mariat C, Afiani A, Thibaudin L (2011) Predicting the risk for dialysis or death in IgA nephropathy. J Am Soc Nephrol 22(4):752–761. https://doi.org/10.1681/asn.2010040355
Moresco RN, Speeckaert MM, Delanghe JR (2015) Diagnosis and monitoring of IgA nephropathy: the role of biomarkers as an alternative to renal biopsy. Autoimmun Rev 14(10):847–853. https://doi.org/10.1016/j.autrev.2015.05.009
Chang S, Li XK (2020) The role of immune modulation in pathogenesis of IgA nephropathy. Front Med 7:92. https://doi.org/10.3389/fmed.2020.00092
Rajasekaran A, Julian BA, Rizk DV (2021) IgA nephropathy: an interesting autoimmune kidney disease. Am J Med Sci 361(2):176–194. https://doi.org/10.1016/j.amjms.2020.10.003
Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, Wyatt RJ, Scolari F, Mestecky J, Gharavi AG, Julian BA (2011) The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22(10):1795–1803. https://doi.org/10.1681/asn.2011050464
Hassler JR (2020) IgA nephropathy: a brief review. Semin Diagn Pathol 37(3):143–147. https://doi.org/10.1053/j.semdp.2020.03.001
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57–74. https://doi.org/10.1038/nature11247
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10(6):925–933. https://doi.org/10.4161/rna.24604
Zuo N, Li Y, Liu N, Wang L (2017) Differentially expressed long non-coding RNAs and mRNAs in patients with IgA nephropathy. Mol Med Rep 16(5):7724–7730. https://doi.org/10.3892/mmr.2017.7542
Luo ZF, Tang D, Xu HX, Lai LS, Chen JJ, Lin H, Yan Q, Zhang XZ, Wang G, Dai Y, Sui WG (2020) Differential expression of transfer RNA-derived small RNAs in IgA nephropathy. Medicine 99(48):e23437. https://doi.org/10.1097/md.0000000000023437
Liu H, Liu D, Liu Y, Xia M, Li Y, Li M, Liu H (2020) Comprehensive analysis of circRNA expression profiles and circRNA-associated competing endogenous RNA networks in IgA nephropathy. PeerJ 8:e10395. https://doi.org/10.7717/peerj.10395
Wang Y, Wu Q, Wang J, Li L, Sun X, Zhang Z, Zhang L (2020) Co-delivery of p38α MAPK and p65 siRNA by novel liposomal glomerulus-targeting nano carriers for effective immunoglobulin a nephropathy treatment. J Control Release 320:457–468. https://doi.org/10.1016/j.jconrel.2020.01.024
Wang C, Ye M, Zhao Q, Xia M, Liu D, He L, Chen G, Peng Y, Liu H (2019) Loss of the golgi matrix protein 130 cause aberrant IgA1 glycosylation in IgA nephropathy. Am J Nephrol 49(4):307–316. https://doi.org/10.1159/000499110
Makita Y, Suzuki H, Kano T, Takahata A, Julian BA, Novak J, Suzuki Y (2020) TLR9 activation induces aberrant IgA glycosylation via APRIL- and IL-6-mediated pathways in IgA nephropathy. Kidney Int 97(2):340–349. https://doi.org/10.1016/j.kint.2019.08.022
Wang X, Shen E, Wang Y, Li J, Cheng D, Chen Y, Gui D, Wang N (2016) Cross talk between miR-214 and PTEN attenuates glomerular hypertrophy under diabetic conditions. Sci Rep 6:31506. https://doi.org/10.1038/srep31506
Bao H, Chen H, Zhu X, Zhang M, Yao G, Yu Y, Qin W, Zeng C, Zen K, Liu Z (2014) MiR-223 downregulation promotes glomerular endothelial cell activation by upregulating importin α4 and α5 in IgA nephropathy. Kidney Int 85(3):624–635. https://doi.org/10.1038/ki.2013.469
Bao H, Hu S, Zhang C, Shi S, Qin W, Zeng C, Zen K, Liu Z (2014) Inhibition of miRNA-21 prevents fibrogenic activation in podocytes and tubular cells in IgA nephropathy. Biochem Biophys Res Commun 444(4):455–460. https://doi.org/10.1016/j.bbrc.2014.01.065
Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC (2011) Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers 30(4):171–179. https://doi.org/10.3233/dma-2011-0766
Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC (2010) Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Investig 90(1):98–103. https://doi.org/10.1038/labinvest.2009.118
Wen L, Zhao Z, Xiao J, Wang Z, He X, Birn H (2018) Renal miR-148b is associated with megalin down-regulation in IgA nephropathy. Biosci Rep. https://doi.org/10.1042/bsr20181578
Li C, Shi J, Zhao Y (2018) MiR-320 promotes B cell proliferation and the production of aberrant glycosylated IgA1 in IgA nephropathy. J Cell Biochem 119(6):4607–4614. https://doi.org/10.1002/jcb.26628
Liu D, Xia M, Liu Y, Tan X, He L, Liu Y, Chen G, Liu H (2020) The upregulation of miR-98-5p affects the glycosylation of IgA1 through cytokines in IgA nephropathy. Int Immunopharmacol 82:106362. https://doi.org/10.1016/j.intimp.2020.106362
Li Y, Xia M, Peng L, Liu H, Chen G, Wang C, Yuan D, Liu Y, Liu H (2021) Downregulation of miR-214-3p attenuates mesangial hypercellularity by targeting PTEN-mediated JNK/c-Jun signaling in IgA nephropathy. Int J Biol Sci 17(13):3343–3355. https://doi.org/10.7150/ijbs.61274
Hennino MF, Buob D, Van der Hauwaert C, Gnemmi V, Jomaa Z, Pottier N, Savary G, Drumez E, Noël C, Cauffiez C, Glowacki F (2016) miR-21-5p renal expression is associated with fibrosis and renal survival in patients with IgA nephropathy. Sci Rep 6:27209. https://doi.org/10.1038/srep27209
Rudnicki M, Perco P, Barbara DH, Leierer J, Heinzel A, Mühlberger I, Schweibert N, Sunzenauer J, Regele H, Kronbichler A (2016) Renal microRNA- and RNA-profiles in progressive chronic kidney disease. Eur J Clin Investig 46(3):213–226. https://doi.org/10.1111/eci.12585
Liang Y, Zhao G, Tang L, Zhang J, Li T, Liu Z (2016) MiR-100-3p and miR-877-3p regulate overproduction of IL-8 and IL-1β in mesangial cells activated by secretory IgA from IgA nephropathy patients. Exp Cell Res 347(2):312–321. https://doi.org/10.1016/j.yexcr.2016.08.011
Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y (2008) Microarray analysis of micro-ribonucleic acid expression in primary immunoglobulin A nephropathy. Saudi Med J 29(10):1388–1393
Pawluczyk IZA, Didangelos A, Barbour SJ, Er L, Becker JU, Martin R, Taylor S, Bhachu JS, Lyons EG, Jenkins RH, Fraser D, Molyneux K, Perales-Patón J, Saez-Rodriguez J, Barratt J (2021) Differential expression of microRNA miR-150-5p in IgA nephropathy as a potential mediator and marker of disease progression. Kidney Int 99(5):1127–1139. https://doi.org/10.1016/j.kint.2020.12.028
Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R (2007) MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 104(9):3432–3437. https://doi.org/10.1073/pnas.0611192104
Ruszkowski J, Lisowska KA, Pindel M, Heleniak Z, Dębska-Ślizień A, Witkowski JM (2019) T cells in IgA nephropathy: role in pathogenesis, clinical significance and potential therapeutic target. Clin Exp Nephrol 23(3):291–303. https://doi.org/10.1007/s10157-018-1665-0
Serino G, Sallustio F, Cox SN, Pesce F, Schena FP (2012) Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol 23(5):814–824. https://doi.org/10.1681/asn.2011060567
Cox SN, Sallustio F, Serino G, Pontrelli P, Verrienti R, Pesce F, Torres DD, Ancona N, Stifanelli P, Zaza G, Schena FP (2010) Altered modulation of WNT-beta-catenin and PI3K/Akt pathways in IgA nephropathy. Kidney Int 78(4):396–407. https://doi.org/10.1038/ki.2010.138
Serino G, Sallustio F, Curci C, Cox SN, Pesce F, De Palma G, Schena FP (2015) Role of let-7b in the regulation of N-acetylgalactosaminyltransferase 2 in IgA nephropathy. Nephrol Dial Transpl 30(7):1132–1139. https://doi.org/10.1093/ndt/gfv032
Serino G, Pesce F, Sallustio F, De Palma G, Cox SN, Curci C, Zaza G, Lai KN, Leung JC, Tang SC, Papagianni A, Stangou M, Goumenos D, Gerolymos M, Takahashi K, Yuzawa Y, Maruyama S, Imai E, Schena FP (2016) In a retrospective international study, circulating miR-148b and let-7b were found to be serum markers for detecting primary IgA nephropathy. Kidney Int 89(3):683–692. https://doi.org/10.1038/ki.2015.333
Zhai Y, Qi Y, Long X, Dou Y, Liu D, Cheng G, Xiao J, Liu Z, Zhao Z (2019) Elevated hsa-miR-590-3p expression down-regulates HMGB2 expression and contributes to the severity of IgA nephropathy. J Cell Mol Med 23(11):7299–7309. https://doi.org/10.1111/jcmm.14582
Xu BY, Meng SJ, Shi SF, Liu LJ, Lv JC, Zhu L, Zhang H (2020) MicroRNA-21-5p participates in IgA nephropathy by driving T helper cell polarization. J Nephrol 33(3):551–560. https://doi.org/10.1007/s40620-019-00682-3
Wang Z, Liao Y, Wang L, Lin Y, Ye Z, Zeng X, Liu X, Wei F, Yang N (2020) Small RNA deep sequencing reveals novel miRNAs in peripheral blood mononuclear cells from patients with IgA nephropathy. Mol Med Rep 22(4):3378–3386. https://doi.org/10.3892/mmr.2020.11405
Yang X, Wu D, Du H, Nie F, Pang X, Xu Y (2017) MicroRNA-135a is involved in podocyte injury in a transient receptor potential channel 1-dependent manner. Int J Mol Med 40(5):1511–1519. https://doi.org/10.3892/ijmm.2017.3152
Jin LW, Ye HY, Xu XY, Zheng Y, Chen Y (2018) MiR-133a/133b inhibits Treg differentiation in IgA nephropathy through targeting FOXP3. Biomed Pharmacother 101:195–200. https://doi.org/10.1016/j.biopha.2018.02.022
Shen M, Pan X, Gao Y, Ye H, Zhang J, Chen Y, Pan M, Huang W, Xu X, Zhao Y, Jin L (2021) LncRNA CRNDE exacerbates IgA nephropathy progression by promoting NLRP3 inflammasome activation in macrophages. Immunol Investig. https://doi.org/10.1080/08820139.2021.1989461
Hu S, Bao H, Xu X, Zhou X, Qin W, Zeng C, Liu Z (2015) Increased miR-374b promotes cell proliferation and the production of aberrant glycosylated IgA1 in B cells of IgA nephropathy. FEBS Lett 589(24):4019–4025. https://doi.org/10.1016/j.febslet.2015.10.033
Yang L, Zhang X, Peng W, Wei M, Qin W (2017) MicroRNA-155-induced T lymphocyte subgroup drifting in IgA nephropathy. Int Urol Nephrol 49(2):353–361. https://doi.org/10.1007/s11255-016-1444-3
Duan ZY, Cai GY, Li JJ, Bu R, Chen XM (2017) Urinary erythrocyte-derived miRNAs: emerging role in IgA nephropathy. Kidney Blood Press Res 42(4):738–748. https://doi.org/10.1159/000481970
Wu J, Zhang H, Wang W, Zhu M, Qi LW, Wang T, Cheng W, Zhu J, Shan X, Huang Z, Zhang L, Chen Y, Sun B, Zhao X, Qian J, Zhu W, Zhou X, Xing C (2018) Plasma microRNA signature of patients with IgA nephropathy. Gene 649:80–86. https://doi.org/10.1016/j.gene.2018.01.050
Kouri NM, Stangou M, Lioulios G, Mitsoglou Z, Serino G, Chiurlia S, Cox SN, Stropou P, Schena FP, Papagianni A (2021) Serum levels of miR-148b and Let-7b at diagnosis may have important impact in the response to treatment and long-term outcome in IgA nephropathy. J Clin Med. https://doi.org/10.3390/jcm10091987
Hu H, Wan Q, Li T, Qi D, Dong X, Xu Y, Chen H, Liu H, Huang H, Wei C, Zhou W, Jiang S, Mo Z, Liao F, Xu Q, He Y (2020) Circulating MiR-29a, possible use as a biomarker for monitoring IgA nephropathy. Iran J Kidney Dis 14(2):107–118
Szeto CC, Wang G, Ng JK, Kwan BC, Mac-Moune Lai F, Chow KM, Luk CC, Lai KB, Li PK (2019) Urinary miRNA profile for the diagnosis of IgA nephropathy. BMC Nephrol 20(1):77. https://doi.org/10.1186/s12882-019-1267-4
Luan R, Tian G, Ci X, Zheng Q, Wu L, Lu X (2021) Differential expression analysis of urinary exosomal circular RNAs in patients with IgA nephropathy. Nephrology (Carlton) 26(5):432–441. https://doi.org/10.1111/nep.13855
Luan R, Tian G, Zhang H, Shi X, Li J, Zhang R, Lu X (2021) Urinary exosomal circular RNAs of sex chromosome origin are associated with gender-related risk differences of clinicopathological features in patients with IgA nephropathy. J Nephrol. https://doi.org/10.1007/s40620-021-01118-7
Duan ZY, Cai GY, Bu R, Lu Y, Hou K, Chen XM (2016) Selection of urinary sediment miRNAs as specific biomarkers of IgA nephropathy. Sci Rep 6:23498. https://doi.org/10.1038/srep23498
Wang G, Kwan BC, Lai FM, Chow KM, Kam-Tao Li P, Szeto CC (2010) Expression of microRNAs in the urinary sediment of patients with IgA nephropathy. Dis Markers 28(2):79–86. https://doi.org/10.3233/dma-2010-0687
Tan K, Chen J, Li W, Chen Y, Sui W, Zhang Y, Dai Y (2013) Genome-wide analysis of microRNAs expression profiling in patients with primary IgA nephropathy. Genome 56(3):161–169. https://doi.org/10.1139/gen-2012-0159
Szeto CC, Ching-Ha KB, Ka-Bik L, Mac-Moune LF, Cheung-Lung CP, Gang W, Kai-Ming C, Kam-Tao LP (2012) Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis Markers 33(3):137–144. https://doi.org/10.1155/2012/842764
Trionfini P, Benigni A (2017) MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol 28(6):1686–1696. https://doi.org/10.1681/asn.2016101117
Liu L, Duan A, Guo Q, Sun G, Cui W, Lu X, Yu H, Luo P (2021) Detection of microRNA-33a-5p in serum, urine and renal tissue of patients with IgA nephropathy. Exp Ther Med 21(3):205. https://doi.org/10.3892/etm.2021.9638
Min QH, Chen XM, Zou YQ, Zhang J, Li J, Wang Y, Li SQ, Gao QF, Sun F, Liu J, Xu YM, Lin J, Huang LF, Huang B (2018) Differential expression of urinary exosomal microRNAs in IgA nephropathy. J Clin Lab Anal. https://doi.org/10.1002/jcla.22226
Szeto CC, Ng JK, Fung WW, Chan GC, Luk CC, Lai KB, Wang G, Chow KM, Mac-Moune Lai F (2022) Urinary mi-106a for the diagnosis of IgA nephropathy: Liquid biopsy for kidney disease. Clin Chim Acta 530:81–86. https://doi.org/10.1016/j.cca.2022.03.006
Xiao B, Wang LN, Li W, Gong L, Yu T, Zuo QF, Zhao HW, Zou QM (2018) Plasma microRNA panel is a novel biomarker for focal segmental glomerulosclerosis and associated with podocyte apoptosis. Cell Death Dis 9(5):533. https://doi.org/10.1038/s41419-018-0569-y
Wang N, Bu R, Duan Z, Zhang X, Chen P, Li Z, Wu J, Cai G, Chen X (2015) Profiling and initial validation of urinary microRNAs as biomarkers in IgA nephropathy. PeerJ 3:e990. https://doi.org/10.7717/peerj.990
Liang S, Cai GY, Duan ZY, Liu SW, Wu J, Lv Y, Hou K, Li ZX, Zhang XG, Chen XM (2017) Urinary sediment miRNAs reflect tubulointerstitial damage and therapeutic response in IgA nephropathy. BMC Nephrol 18(1):63. https://doi.org/10.1186/s12882-017-0482-0
Duan A, Liu L, Lou Y, Zhang D, Li H, Chen Y, Cui W, Miao L (2019) Diagnostic value of urinary miR-152–5p in patients with IgA nephropathy with elevated proteinuria levels. Clin Lab. https://doi.org/10.7754/Clin.Lab.2019.190111
Mbemi A, Khanna S, Njiki S, Yedjou CG (2020) Impact of gene-environment interactions on cancer development. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph17218089
Sanchez-Rodriguez E, Southard CT, Kiryluk K (2021) GWAS-based discoveries in IgA nephropathy, membranous nephropathy, and steroid-sensitive nephrotic syndrome. Clin J Am Soc Nephrol 16(3):458–466. https://doi.org/10.2215/cjn.14031119
Zhong Z, Feng S, Shi D, Xu R, Yin P, Wang M, Mao H, Huang F, Li Z, Yu X, Li M (2019) Association of FCRL3 gene polymorphisms with IgA nephropathy in a Chinese Han population. DNA Cell Biol 38(10):1155–1165. https://doi.org/10.1089/dna.2019.4900
Feng Y, Su Y, Ma C, Jing Z, Yang X, Zhang D, Xie M, Li W, Wei J (2019) 3’UTR variants of TNS3, PHLDB1, NTN4, and GNG2 genes are associated with IgA nephropathy risk in Chinese Han population. Int Immunopharmacol 71:295–300. https://doi.org/10.1016/j.intimp.2019.03.041
Yamada K, Huang ZQ, Raska M, Reily C, Anderson JC, Suzuki H, Kiryluk K, Gharavi AG, Julian BA, Willey CD, Novak J (2020) Leukemia inhibitory factor signaling enhances production of galactose-deficient IgA1 in IgA nephropathy. Kidney Dis (Basel, Switzerland) 6(3):168–180. https://doi.org/10.1159/000505748
Yang K, Zhang Y, Mai X, Hu L, Ma C, Wei J (2021) MIR17HG genetic variations affect the susceptibility of IgA nephropathy in Chinese Han people. Gene 800:145838. https://doi.org/10.1016/j.gene.2021.145838
Shajari N, Mansoori B, Davudian S, Mohammadi A, Baradaran B (2017) Overcoming the challenges of siRNA delivery: nanoparticle strategies. Curr Drug Deliv 14(1):36–46. https://doi.org/10.2174/1567201813666160816105408
Wu Y, Huang Q, Wang J, Dai Y, Xiao M, Li Y, Zhang H, Xiao W (2021) The feasibility of targeted magnetic iron oxide nanoagent for noninvasive IgA nephropathy diagnosis. Front Bioeng Biotechnol 9:755692. https://doi.org/10.3389/fbioe.2021.755692
Zhang X, Wu Y, Sun K, Tan J (2014) Effect of erythropoietin loading chitosan-tripolyphosphate nanoparticles on an IgA nephropathy rat model. Exp Ther Med 7(6):1659–1662. https://doi.org/10.3892/etm.2014.1643
Acknowledgements
The authors are grateful to the authorities of Aarupadai Veedu Medical College and Hospital, Vinayaka Missions Research Foundation (Deemed to be University), and we would like to thank BioRender (https://biorender.com) used to create figures.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to content, writing, reviewing and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human and animal participation performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kademani, S.P., Nelaturi, P., Sathyasagar, K. et al. Noncoding RNAs associated with IgA nephropathy. J Nephrol 36, 911–923 (2023). https://doi.org/10.1007/s40620-022-01498-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40620-022-01498-4