Abstract
Purpose of Review
RNA-sequencing has provided a new way to study transcriptomic population differences of non-model organisms such as marine invertebrates. Importantly, population transcriptomics can provide insight regarding the genetic mechanisms that allow some populations to tolerate various environmental stressors in this era of climate change. Such studies can help us predict future population dynamics and species survival for marine invertebrates of ecological and economic value. Here, I review practical considerations for marine invertebrate researchers using RNA-sequencing, recent applications of population transcriptomics in marine invertebrates, current limitations of this approach, and future areas of study.
Recent Findings
Recent marine invertebrate studies have examined population-specific responses to abiotic stressors including temperature, salinity, pH, and water quality. Populations tolerant of environmental stressors generally show high constitutive gene expression before stress exposure and a muted overall transcriptional response following stress exposure. Evidence for molecular signatures of selection on several genes including ATP synthase and aquaporins has been identified in marine invertebrate populations exposed to broad temperature regimes. At present, the genetic mechanisms underlying individual variation within a population and the transcriptome response to multiple environmental stressors remain poorly understood.
Summary
Complete assessment of marine invertebrate population responses to climate change will require pairing RNA-sequencing approaches with complementary phenotype studies. Improvements in transcriptome annotation and future work investigating gene expression regulation will lead to an increased understanding of the genes responsible for population-level tolerance to environmental stressors.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol. 2017;13(5):23. https://doi.org/10.1371/journal.pcbi.1005457.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. https://doi.org/10.1038/nrg2484.
Schoville SD, Barreto FS, Moy GW, Wolff A, Burton RS. Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol Biol. 2012;12:17. https://doi.org/10.1186/1471-2148-12-170.
Wang YJ. The future of marine invertebrates in face of global climate change. J Coast Dev. 2014;17(1):e105. https://doi.org/10.4172/1410-5217.1000e105.
Prather CM, Pelini SL, Laws A, Rivest E, Woltz M, Bloch CP, et al. Invertebrates, ecosystem services and climate change. Biol Rev. 2013;88(2):327–48. https://doi.org/10.1111/brv.12002.
Ibañez E, Herrero M, Mendiola JA, Castro-Puyana M. Extraction and characterization of bioactive compounds with health benefits from marine resources: macro and micro algae, cyanobacteria, and invertebrates. In: Hayes M, editor. Marine bioactive compounds. Boston: Springer; 2012.
Lopez JV, Kamel B, Medina M, Collins T, Baums IB. Multiple Facets of Marine Invertebrate Conservation Genomics. Annu Rev Anim Biosci. 2019;7(1). https://doi.org/10.1146/annurev-animal-020518-115034.
Alvarez M, Schrey AW, Richards CL. Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Mol Ecol. 2015;24(4):710–25. https://doi.org/10.1111/mec.13055.
DeBiasse MB, Kelly MW. Plastic and evolved responses to global change: what can we learn from comparative transcriptomics? J Hered. 2016;107(1):71–81. https://doi.org/10.1093/jhered/esv073.
Jax E, Wink M, Kraus RHS. Avian transcriptomics: opportunities and challenges. J Ornithol. 2018;159(3):599–629. https://doi.org/10.1007/s10336-018-1532-5.
Matz MV. Fantastic beasts and how to sequence them: ecological genomics for obscure model organisms. Trends Genet. 2018;34(2):121–32. https://doi.org/10.1016/j.tig.2017.11.002.
Marshall KED EJ, Petrunina A, Kolbasov G, Chan BKK. Transcriptional dynamics following freezing stress reveal selection for mechanisms of freeze tolerance at the poleward range margin in the cold water intertidal barnacle Semibalanus balanoides. bioRxiv. 2018:449330. https://doi.org/10.1101/449330.
Maynard A, Bible JM, Pespeni MH, Sanford E, Evans TG. Transcriptomic responses to extreme low salinity among locally adapted populations of Olympia oyster (Ostrea lurida). Mol Ecol. 2018;27(21):4225–40. https://doi.org/10.1111/mec.14863.
• Kirk NL, Howells EJ, Abrego D, Burt JA, Meyer E. Genomic and transcriptomic signals of thermal tolerance in heat-tolerant corals (Platygyra daedalea) of the Arabian/Persian gulf. Mol Ecol. 2018;27(24):5180–94. https://doi.org/10.1111/mec.14934. This paper found that heat tolerance in Platygyra corals is affected by both nucleotide sequences and differential gene expression.
Clark MS, Thorne MAS, King M, Hipperson H, Hoffman JI, Peck LS. Life in the intertidal: cellular responses, methylation and epigenetics. Funct Ecol. 2018;32(8):1982–94. https://doi.org/10.1111/1365-2435.13077.
•• DeBiasse MB, Kawji Y, Kelly MW. Phenotypic and transcriptomic responses to salinity stress across genetically and geographically divergent Tigriopus californicus populations. Mol Ecol. 2018;27(7):1621–32. https://doi.org/10.1111/mec.14547. This work investigated salinity response in intertidal copepods and illustrates the importance of creating separate transcriptome assemblies for each population and of using additional phenotype and post-transcriptional/post-translational approaches to complement RNA-seq.
•• Feis ME, John U, Lokmer A, Luttikhuizen PC, Wegner KM. Dual transcriptomics reveals co-evolutionary mechanisms of intestinal parasite infections in blue mussels Mytilus edulis. Mol Ecol. 2018;27(6):1505–19. https://doi.org/10.1111/mec.14541. This paper discusses strategies for circumventing low annotation rates of marine invertebrate de novo transcriptome assemblies.
Schuierer S, Carbone W, Knehr J, Petitjean V, Fernandez A, Sultan M, et al. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics. 2017;18:13. https://doi.org/10.1186/s12864-017-3827-y.
Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:96.
Denny MW. Invertebrate mucous secretions: functional alternatives to vertebrate paradigms. Symp Soc Exp Biol. 1989;43:337–66.
Denny MW. Molecular biomechanics of molluscan mucous secretions. In: Wilbur KS, Hochachka PW, editors. The Mollusca. New York: Academic Press; 1983. p. 431–65.
Smith AM. The structure and function of adhesive gels from invertebrates. Integr Comp Biol. 2002;42(6):1164–71. https://doi.org/10.1093/icb/42.6.1164.
Gleason LU, Burton RS. RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Mol Ecol. 2015;24(3):610–27. https://doi.org/10.1111/mec.13047.
Lee JTY, Cheung KMC, Leung VYL. Extraction of RNA from tough tissues with high proteoglycan content by cryosection, second phase separation and high salt precipitation. J Biol Methods. 2015;2(2):e20.
Le Bleu HK, Kamal FA, Kelly M, Ketz JP, Zuscik MJ, Elbarbary RA. Extraction of high-quality RNA from human articular cartilage. Anal Biochem. 2017;518:134–8. https://doi.org/10.1016/j.ab.2016.11.018.
Grosberg RKC, C. W. Genetic structure in the sea: from populations to communities. In: Bertness MDG, Gaines SD, Hay ME, editors. Marine community ecology. Sunderland: Sinauer; 2001. p. 61–84.
Weersing K, Toonen RJ. Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser. 2009;393:1–12. https://doi.org/10.3354/meps08287.
Morgan SG, Fisher JL, Miller SH, McAfee ST, Largier JL. Nearshore larval retention in a region of strong upwelling and recruitment limitation. Ecology. 2009;90(12):3489–502. https://doi.org/10.1890/08-1550.1.
Drake PT, Edwards CA, Morgan SG, Dever EP. Influence of larval behavior on transport and population connectivity in a realistic simulation of the California current system. J Mar Res. 2013;71(4):317–50. https://doi.org/10.1357/002224013808877099.
Sanford E, Kelly MW. Local adaptation in marine invertebrates. Annu Rev Mar Sci. 2011;3:509–35. https://doi.org/10.1146/annurev-marine-120709-142756.
Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12(10):671–82. https://doi.org/10.1038/nrg3068.
Graham AM, Barreto FS. Novel microRNAs are associated with population divergence in transcriptional response to thermal stress in an intertidal copepod. Mol Ecol. 2019;28(3):584–99. https://doi.org/10.1111/mec.14973.
Griffiths JS, Pan FT-C, Kelly MW. Differential responses to ocean acidification between populations of Balanophyllia elegans corals from high and low upwelling environments. Mol Ecol. 2019. https://doi.org/10.1111/mec.15050.
• Rocker MM, Kenkel CD, Francis DS, Willis BL, Bay LK. Plasticity in gene expression and fatty acid profiles of Acropora tenuis reciprocally transplanted between two water quality regimes in the central Great Barrier Reef, Australia. J Exp Mar Biol Ecol. 2019;511:40–53. https://doi.org/10.1016/j.jembe.2018.11.004. This paper found a stronger effect of the source population (e.g. adaptation) compared to the transplant location (e.g. plasticity) on gene expression in Acropora corals.
Chen N, Huang ZK, Lu CK, Shen YW, Luo X, Ke CH, et al. Different transcriptomic responses to thermal stress in heat-tolerant and heat-sensitive Pacific abalones indicated by cardiac performance. Front Physiol. 2019;9:14. https://doi.org/10.3389/fphys.2018.01895.
• Rose NH, Bay RA, Morikawa MK, Palumbi SR. Polygenic evolution drives species divergence and climate adaptation in corals. Evolution. 2018;72(1):82–94. https://doi.org/10.1111/evo.13385. This work illustrates that the early stages of speciation in Acropora corals likely results from polymorphic alleles across many loci that affect gene expression.
Pante E, Becquet V, Viricel A, Garcia P. Investigation of the molecular signatures of selection on ATP synthase genes in the marine bivalve Limecola balthica. Aquat Living Resour. 2019;32:7. https://doi.org/10.1051/alr/2019001.
Dong ZG, Zhang DD, Li XY, Lu X, Chen L, Niu DH, et al. Twenty-nine SNP markers developed from the transcriptomics of rainbow clam Moerella iridescens and their application in population genetics. Conserv Genet Resour. 2018;10(3):277–9. https://doi.org/10.1007/s12686-017-0801-6.
ChinBA. Characterizing the role of DNA methylation patterns in the California mussel, Mytilus californianus: Sonoma State University; 2018.
Barshis DJ, Ladner JT, Oliver TA, Seneca FO, Traylor-Knowles N, Palumbi SR. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci U S A. 2013;110(4):1387–92. https://doi.org/10.1073/pnas.1210224110.
Fabbri EV, Franzellitti S. HSP expression in bivalves. ISJ. 2008;5:135–61.
Tomanek L. Proteomics to study adaptations in marine organisms to environmental stress. J Proteome. 2014;105:92–106. https://doi.org/10.1016/j.jprot.2014.04.009.
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2018;36:3420–35. https://doi.org/10.1093/nar/gkn176.
Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–93. https://doi.org/10.1093/bioinformatics/bti430.
Jonas K, Liu J, Chien P, Laub MT. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154(3):623–36. https://doi.org/10.1016/j.cell.2013.06.034.
Kenkel CD, Meyer E, Matz MV. Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol Ecol. 2013;22(16):4322–34. https://doi.org/10.1111/mec.12390.
Somero GN, Lockwood BL, Tomanek L. Biochemical adaptation: response to environmental challenges, from Life's origins to the Anthropocene. MA:Sinauer Associates: Sunderland; 2017.
DoneySC, FabryVJ, FeelyRA, Kleypas JA. Ocean Acidification: The Other CO2 Problem. Annual Review of Marine Science. Annual Review of Marine Science. Palo Alto: Annual Reviews; 2009. p. 169–192.
Feely RA, Sabine CL, Hernandez-Ayon JM, Ianson D, Hales B. Evidence for upwelling of corrosive "acidified" water onto the continental shelf. Science. 2008;320(5882):1490–2. https://doi.org/10.1126/science.1155676.
Rocker MM, Francis DS, Fabricius KE, Willis BL, Bay LK. Variation in the health and biochemical condition of the coral Acropora tenuis along two water quality gradients on the great barrier reef, Australia. Mar Pollut Bull. 2017;119:106–19. https://doi.org/10.1016/j.marpolbul.2017.03.066.
Dixon GB, Davies SW, Aglyamova GV, Meyer E, Bay LK, Matz MV. Genomic determinants of coral heat tolerance across latitudes. Science. 2015;348:1460–2. https://doi.org/10.1126/science.1261224.
Greenwood JM, Ezquerra AL, Behrens S, Branca A, Mallet L. Current analysis of host-parasite interactions with a focus on next generation sequencing data. Zoology. 2016;119(4):298–306. https://doi.org/10.1016/j.zool.2016.06.010.
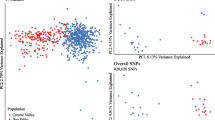
De Wit P, Pespeni MH, Palumbi SR. SNP genotyping and population genomics from expressed sequences - current advances and future possibilities. Mol Ecol. 2015;24(10):2310–23. https://doi.org/10.1111/mec.13165.
Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–16. https://doi.org/10.1038/nrg2063.
Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y. The genetic architecture of gene expression levels in wild baboons. eLife. 2015;4:22. https://doi.org/10.7554/eLife.04729.
Philip BN, Yi SX, Elnitsky MA, Lee RE. Aquaporins play a role in desiccation and freeze tolerance in larvae of the goldenrod gall fly, Eurosta solidaginis. J Exp Biol. 2008;211(7):1114–9. https://doi.org/10.1242/jeb.016758.
Fangue NA, Hofmeister M, Schulte PM. Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish, Fundulus heteroclitus. J Exp Biol. 2006;209(15):2859–72. https://doi.org/10.1242/jeb.02260.
Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. https://doi.org/10.1146/annurev.physiol.61.1.243.
Zhang GF, Fang XD, Guo XM, Li L, Luo RB, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490(7418):49–54. https://doi.org/10.1038/nature11413.
McAfee D, Cumbo VR, Bishop MJ, Raftos DA. Intraspecific differences in the transcriptional stress response of two populations of Sydney rock oyster increase with rising temperatures. Mar Ecol Prog Ser. 2018;589:115–27. https://doi.org/10.3354/meps12455.
Gleason LU, Burton RS. Phenotypic evidence for local adaptation to heat stress in the marine snail Chlorostoma (formerly Tegula) funebralis. J Exp Mar Biol Ecol. 2013;448:360–6. https://doi.org/10.1016/j.jembe.2013.08.008.
Day DAT, M. F. Post-transcriptional gene regulatory mechanisms in eukaruotes: an overview. J Endocrinol. 1998;157(3):361–71.
Kia-Ki HM, A. Post-translational chemical modification(s) of proteins. Int J Biochem. 1992;24(1):19–28. https://doi.org/10.1016/0020-711X(92)90225-P.
Lockwood BL, Somero GN. Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol Ecol. 2011;20(3):517–29. https://doi.org/10.1111/j.1365-294X.2010.04973.x.
Willett CS, Burton RS. Proline biosynthesis genes and their regulation under salinity stress in the euryhaline copepod Tigriopus californicus. Comp Biochem Physiol B Biochem Mol Biol. 2002;132(4):739–50. https://doi.org/10.1016/s1096-4959(02)00091-x.
Romero MR, Perez-Figueroa A, Carrera M, Swanson WJ, Skibinski DOF, Diz AP. RNA-seq coupled to proteomic analysis reveals high sperm proteome variation between two closely related marine mussel species. J Proteome. 2019;192:169–87. https://doi.org/10.1016/j.jprot.2018.08.020.
Gallardo-Escarate C, Valenzuela-Munoz V, Nunez-Acuna G. RNA-Seq analysis using De novo transcriptome assembly as a reference for the Salmon louse Caligus rogercresseyi. PLoS One. 2014;9(4):17. https://doi.org/10.1371/journal.pone.0092239.
Yarra T, Gharbi K, Blaxter M, Peck LS, Clark MS. Characterization of the mantle transcriptome in bivalves: Pecten maximus, Mytilus edulis and Crassostrea gigas. Mar Genomics. 2016;27:9–15. https://doi.org/10.1016/j.margen.2016.04.003.
Primmer CR, Papakostas S, Leder EH, Davis MJ, Ragan MA. Annotated genes and nonannotated genomes: cross-species use of gene ontology in ecology and evolution research. Mol Ecol. 2013;22(12):3216–41. https://doi.org/10.1111/mec.12309.
Stanford BCM, Rogers SM. R(NA)-tistic expression: the art of matching unknown mRNA and proteins to environmental response in ecological genomics. Mol Ecol. 2018;27(4):827–30. https://doi.org/10.1111/mec.14419.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:13. https://doi.org/10.1186/1471-2105-9-559.
Plachetzki DC, Pankey MS, Johnson BR, Ronne EJ, Kopp A, Grosberg RK. Gene co-expression modules underlying polymorphic and monomorphic zooids in the colonial hydrozoan, Hydractinia symbiolongicarpus. Integr Comp Biol. 2014;54(2):276–83. https://doi.org/10.1093/icb/icu080.
Rose NH, Seneca FO, Palumbi SR. Gene networks in the wild: identifying transcriptional modules that mediate coral resistance to experimental heat stress. Genome Biol Evol. 2016;8(1):243–52. https://doi.org/10.1093/gbe/evv258.
Bay RA, Palumbi SR. Transcriptome predictors of coral survival and growth in a highly variable environment. Ecol Evol. 2017;7(13):4794–803. https://doi.org/10.1002/ece3.2685.
Wong JM, Johnson KM, Kelly MW, Hofmann G. Transcriptomics reveal transgenerational effects in purple sea urchin embryos: adult acclimation to upwelling conditions alters the response of their progeny to differential pCO2 levels. Mol Ecol. 2018;27(5):1120–37. https://doi.org/10.1111/mec.14503.
Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–4. https://doi.org/10.1093/nar/28.18.3442.
von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–61. https://doi.org/10.1093/nar/gkg034.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. https://doi.org/10.1038/nprot.2015.053.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. https://doi.org/10.1038/nprot.2013.084.
Jones P, Binns D, Chang HY, Fraser M, Li WZ, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. https://doi.org/10.1093/bioinformatics/btu031.
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. https://doi.org/10.1093/nar/25.17.3389.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. https://doi.org/10.1016/j.cell.2009.01.002.
Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12(12):846–60. https://doi.org/10.1038/nrg3079.
Cork JM, Purugganan MD. The evolution of molecular genetic pathways and networks. Bioessays. 2004;26(5):479–84. https://doi.org/10.1002/bies.20026.
Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett. 2008;11(2):106–15. https://doi.org/10.1111/j.1461-0248.2007.01130.x.
Suarez-Ulloa V, Gonzalez-Romero R, Eirin-Lopez JM. Environmental epigenetics: A promising venue for developing next-generation pollution biomonitoring tools in marine invertebrates. Mar Pollut Bull. 2015;98(1–2):5–13. https://doi.org/10.1016/j.marpolbul.2015.06.020.
Hofmann GE. Ecological Epigenetics in Marine Metazoans. Front Mar Sci. 2017;4:7. https://doi.org/10.3389/fmars.2017.00004.
Gavery MR, Roberts SB. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. PeerJ. 2013;1:15. https://doi.org/10.7717/peerj.215.
Denny MW, Dowd WW, Bilir L, Mach KJ. Spreading the risk: small-scale body temperature variation among intertidal organisms and its implications for species persistence. J Exp Mar Biol Ecol. 2011;400(1–2):175–90. https://doi.org/10.1016/j.jembe.2011.02.006.
Jimenez AG, Jayawardene S, Alves S, Dallmer J, Dowd WW. Micro-scale environmental variation amplifies physiological variation among individual mussels. Proc R Soc B Biol Sci. 2015;282(1820). https://doi.org/10.1098/rspb.2015.2273.
Gleason LU, Miller LP, Winnikoff JR, Somero GN, Yancey PH, Bratz D, et al. Thermal history and gape of individual Mytilus californianus correlate with oxidative damage and thermoprotective osmolytes. J Exp Biol. 2017;220(22):4292–304. https://doi.org/10.1242/jeb.168450.
Xu Q, Zhu CY, Fan YY, Song ZH, Xing SL, Liu W, et al. Population transcriptomics uncovers the regulation of gene expression variation in adaptation to changing environment. Sci Rep. 2016;6:10. https://doi.org/10.1038/srep25536.
Moya A, Ganot P, Furla P, Sabourault C. The transcriptomic response to thermal stress is immediate, transient and potentiated by ultraviolet radiation in the sea anemone Anemonia viridis. Mol Ecol. 2012;21(5):1158–74. https://doi.org/10.1111/j.1365-294X.2012.05458.x.
Padilla-Gamino JL, Kelly MW, Evans TG, Hofmann GE. Temperature and CO2 additively regulate physiology, morphology and genomic responses of larval sea urchins, Strongylocentrotus purpuratus. Proc R Soc B Biol Sci. 2013;280(1759):9. https://doi.org/10.1098/rspb.2013.0155.
Vidal-Dupiol J, Dheilly NM, Rondon R, Grunau C, Cosseau C, Smith KM, et al. Thermal stress triggers broad Pocillopora damicornistranscriptomic remodeling, while Vibrio coralliilyticusinfection induces a more targeted Immuno-suppression response. PLoS One. 2014;9(9):15. https://doi.org/10.1371/journal.pone.0107672.
Mohamed B, Hajer A, Susanna S, Caterina O, Flavio M, Hamadi B, et al. Transcriptomic responses to heat stress and nickel in the mussel Mytilus galloprovincialis. Aquat Toxicol. 2014;148:104–12. https://doi.org/10.1016/j.aquatox.2014.01.004.
Marie AD, Smith S, Green AJ, Rico C, Lejeusne C. Transcriptomic response to thermal and salinity stress in introduced and native sympatric Palaemon caridean shrimps. Sci Rep. 2017;7:12. https://doi.org/10.1038/s41598-017-13631-6.
Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294(5540):151–4. https://doi.org/10.1126/science.1063656.
Duputie A, Massol F, Chuine I, Kirkpatrick M, Ronce O. How do genetic correlations affect species range shifts in a changing environment? Ecol Lett. 2012;15(3):251–9. https://doi.org/10.1111/j.1461-0248.2011.01734.x.
Chevin LM. Genetic constraints on adaptation to a changing environment. Evolution. 2013;67(3):708–21. https://doi.org/10.1111/j.1558-5646.2012.01809.x.
Rogers-Bennett L, Dondanville RF, Moore JD, Vilchis LI. Response of red abalone reproduction to warm water, starvation, and disease stressors: implications of ocean warming. J Shellfish Res. 2010;29(3):599–611. https://doi.org/10.2983/035.029.0308.
Parker LM, Scanes E, O'Connor WA, Coleman RA, Byrne M, Portner HO, et al. Ocean acidification narrows the acute thermal and salinity tolerance of the Sydney rock oyster Saccostrea glomerata. Mar Pollut Bull. 2017;122(1–2):263–71. https://doi.org/10.1016/j.marpolbul.2017.06.052.
Acknowledgments
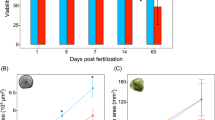
Photographs for Fig.1 were graciously provided by Samantha Clements and Daniel Schneck.
Funding
This study was funded in part by release time provided by the CSU Sacramento College of Natural Sciences and Mathematics.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Lani U. Gleason declares no potential conflict of interest.
Human and Animal Rights and Informed Consent
This article contains no studies with human and animal subjects performed by the author.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Population Genetics
Rights and permissions
About this article
Cite this article
Gleason, L.U. Applications and Future Directions for Population Transcriptomics in Marine Invertebrates. Curr Mol Bio Rep 5, 116–127 (2019). https://doi.org/10.1007/s40610-019-00121-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40610-019-00121-z