Abstract
Purpose of review
Streptococcus suis is an emerging zoonotic pathogen that can cause severe infections in pigs and humans. S. suis has been reported in Europe, America, and Asia, and has resulted in heavy losses to swine production and serious public health concerns. Serum agglutination has identified several serotypes. Here, we review the methods for serotyping S. suis, the current serological classifications, and the future trend in serotyping.
Recent findings
In recent years, some serotypes have been identified as other bacterial species, while others have been identified as novel S. suis serotypes. Modern molecular biology techniques, including new molecular biology serotyping methods based on the serotype-specific genes, are more accurate and sensitive than traditional serotyping methods.
Summary
There are many undiscovered serotypes of S. suis in nature. The relationship between capsule antigenicity and capsular polysaccharide locus genes is complex. Although molecular biology techniques are simple, rapid, and sensitive, the results obtained by these methods should be validated by serological methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus suis (S. suis) is a Gram-positive bacterium that is a significant zoonotic pathogen [1, 2]. S. suis infections are endemic in many countries [3]. Humans can also be infected with S. suis, which can be transmitted via the handling of infected pig carcasses or meat, especially if there are exposed cuts or abrasions on the hands [3]. Human infection can cause meningitis, septicaemia, endocarditis, and/or deafness. The lack of efficient vaccines, sensitive diagnostics, and sufficient knowledge about the epidemiology of the disease hamper control measures.
So far, 33 S. suis serotypes have been identified based on the presence of specific capsular polysaccharides [4, 5]. The capsular polysaccharide structure of some serotypes has been determined [6,7,8, 9•]; serotype 2 is the most pathogenic and prevalent serotype worldwide, and is also a major zoonotic and human pathogen [10]. Other serotypes are also frequently isolated from infected pigs, including serotypes 3, 4, 8, and 9 [11,12,13,14,15]. Recently, some serotypes, including serotypes 20, 22, 26, and 33, were reclassified as other organisms [16, 17]; meanwhile, a novel serotype was also discovered [18••]. Thus, serotype-based classification of S. suis needs to be reevaluated. New molecular biology identification methods have been used for S. suis serotyping; these methods are based on analysis of capsular polysaccharide synthesis gene clusters [19].
Serological classification
Streptococci were classified by Lancefield grouping in the 1930s based on the carbohydrate composition of bacterial cell wall antigens [20]. In the early 1960s, Streptococcus strains from pigs were designated into Lancefield groups R, S, RS, and T by De Moor [21]. Elliott then revealed that groups R and S were actually subgroups of Lancefield’s group D; these strains were classified as a new species, “Streptococcus suis” [22]. Groups S, R, and RS were later reclassified as S. suis serotypes 1, 2, and 1/2, respectively [22, 23]. In 1983, Perch et al. reported six new serotypes (serotypes 3–8) [24]. Kilpper-Balz and Schleifer finally made the formal proposal of the name “S. suis” in 1987 [25]; subsequently, serotypes 9–34 were discovered between 1989 and 1995 [26,27,28]. Based on variations in capsular antigens, 35 serotypes (1–34 and 1/2) were originally assigned to S. suis species. Recently, Okura et al. suggested that 6 serotypes (serotypes 20, 22, 26, and 32–34) should be removed from S. suis taxonomy [29••]. In addition, phylogenetic analyses using 16S rRNA[30], cpn60 [31], sodA, and recN [17] sequences have demonstrated that reference strains of serotypes 20, 22, 26, and 32–34 are clearly distinguishable from the reference strains of the other 29 serotypes (i.e., serotypes1–19, 21, 23–25, 27–31, and 1/2). Thus, there are 29 serotypes that have been identified as S. suis species; however, most research has included serotypes 20, 23, 26, and 33.
The basis of serotyping
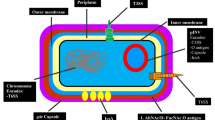
All S. suis serotyping methods are based on either antigenic differences in capsular polysaccharides (CPS) or serotype-specific genes in the capsular polysaccharides synthesis locus (CPS locus). Bacterial CPS are found at the outer surface of the bacterium, where they form an amorphous layer surrounding the cell. They are further organized into a distinct structure termed a capsule and mediate direct interactions between the bacterium and its environment. CPS have been regarded as an important virulence factor for many pathogens [32]. CPS are an incredibly diverse range of antigenic molecules that differ by monosaccharide units as well as how these units are joined together; this is the basis of some serotyping methods [32].
Some S. suis serotypes have one- or two-way cross-reactions that may be due to the similarities in CPS composition (Table 1) [33]. There are complex cross-reactions between serotypes 1/2 and 1, serotypes 1/2 and 2, and serotypes 1 and 14. Cross-absorption experiments showed that serotype 1/2 contains the serotype 2 antigen, and thus all antibody activity against serotype 2 can be removed from anti-serotype 1/2 serum through absorption with serotype 2 germs. The serotype 1 component of serotype 1/2 is not identical to the serotype 1 antigen, as serotype 1/2 strains do not completely exhaust the anti-serotype 1 serum [34]. Serotype 1 strains can react with the serum produced against both serotypes 1 and 14. Antibody activity against serotype 1 can be removed from anti-type 14 serum by absorption with serotype 1 organisms. The adsorbed serum can still agglutinate with serotype 14 strains [26]. The capsular polysaccharide structure of four serotypes was deconvoluted to understand the reason for the complex cross-reaction between the four serotypes (i.e., serotypes 1, 2, 14, and 1/2; Table 2); a sialic acid containing a side chain was found to be the common feature of all four serotypes [9•]. Furthermore, lectin-based studies and genetic analyses of the CPS locus also predicted the presence of this sugar in serotypes 6, 13, 16, and 27 [19, 35]. Although the backbone or side chain is the same in the four serotypes (Table 2), the antigenicity differs between serotypes. The structure and conformation characteristics of the four serotypes are complex. The antigenicity of CPS are decided by the monosaccharide, secondary, and three-dimensional structures [9•]. Additionally, the CPS structure of serotype 9 showed that it is not a member of the S. suis sialylated CPS group (Table 2) [6]. These studies provided the basis for further studies regarding important structural elements recognized by anti-CPS-specific antibodies, which improved our understating of the serological reactions leading to S. suis serotype classification. The relationship between serological characterization and CPS structure requires additional discoveries regarding CPS structure and composition across different serotypes. This will improve S. suis serotyping by serological methods.
CPS are synthesized via a complex pathway; generally, the genes involved in the biosynthesis are clustered in a single locus called the CPS locus. The enzymes to build the repeat unit, including an initial glycosyl phosphate transferase, additional transferases responsible for the formation of the linkages allowing for the addition of sugars (or other moieties) or other modifications of the repeat unit, as well as a repeat-unit flippase and polymerase, are encoded by the CPS locus [32]. The serotype-specific genes in the locus provide the basis for developing molecular serotyping methods. The CPS loci of all S. suis serotypes were sequenced [19, 36, 37]. The length of the CPS locus ranges from 15,274 to 40,198 bp. Most of the CPS genes are oriented in the same direction. In all serotypes, the genes involved in the regulation and processing of capsule (CP), wzg, wzd, wze, and wzh are located on the 5’-side of the locus. All CPS gene clusters also contain genes encoding for putative flippase (wzx) and polymerase (wzy), as well as various sets of glycosyltransferase genes, including an initial sugar transferase gene. This suggests that CPS are synthesized by the wzx/wzy pathway in all S. suis serotypes. Modifying enzymes, nucleotide sugar phosphate biosynthesis enzymes, and other enzymes were predicted to be encoded by some of the CPS genes; these are involved in the biosynthesis of and addition of other components to CPS. Except for the six non-S.suis serotypes (serotypes 20, 22, 26, and 32–34) [29••], most of the CPS loci are located between orfZ and aroA(or glf) on the chromosome and have a cassette-like structure. The serotype-specific genes are flanked by conserved genes common to most gene clusters. This type of CPS cluster is also found in other Streptococcus species [38], including Streptococcus pneumoniae, Streptococcus agalactiae, and Streptococcus thermophilus. The one exception is serotype 27, where the CPS locus is located between the SSU1265 and SSU1264 genes of P1/7. The CPS loci were classified into three groups (i.e., I-a, I-b, and II) according to the location on the chromosome [19]. The percentage of G + C content of all CPS gene loci (32.5 to 36.7%) is lower than that of several S. suis genomes (41.0 to 41.4%). This suggests that these genes may have been imported into S. suis (or their ancestors) on multiple occasions from different and unknown sources; this is supported by the presence of multiple non-homologous or highly divergent forms of key enzymes and horizontal mobile elements (transposases), as well as the lower percentage of G + C content of the region [37].
Serotyping methods
Many S. suis serotypes can infect humans and animals with different epidemiology and pathogenicity [39, 40]. Serotyping is one of the most imperative diagnostic tools for S. suis infections, and remains a valued method for understanding the epidemiology of a specific outbreak or monitoring serotype prevalence, in addition to guiding vaccine development.
Traditional serological serotyping methods
Agglutination and co-agglutination tests are the traditional methods used for serotyping S. suis [33]. In these methods, the isolates are reacted with a panel of anti-sera specific to all S. suis serotypes. It is very arduous and time-consuming, and it is exclusively performed on isolated colonies. Producing anti-sera is also laborious, time-consuming, and expensive. Auto-agglutinating and acapsular strains can not be serotyped using anti-sera methods [33, 41]. Multi-agglutinating isolates are usually detected in animal hosts [42,43,44,45]. Cross-reactions amongst some S. suis serotypes (Table 1) also increases the complexity of serotyping by serological methods. Serological methods cannot readily distinguish between serotypes 2 and 1/2 or serotypes 1 and 14. Thus, serological methods are not suitable for epidemiological investigations with a large number of clinical samples.
Molecular serotyping methods
Polymerase chain reaction (PCR) typing methods provide a quick and economical way to serotype isolates [36, 44]. After the CPS locus of every serotype was sequenced, the serotype-specific genes were identified and used to develop molecular serotyping methods for S. suis. The PCR methods to identify serotypes 1, 2, 7, 9, 14, and 1/2 were developed by Smith et al. in the 1990s [36, 46]. Since the CPS locus sequences of the other serotypes were unknown, there were no PCR method for detecting the other 27 serotypes at that time. Building on the original PCR work, some multiplex PCR methods were reported in the 2000s [47,48,49]. PCR methods were only capable of detecting 15 serotypes until the entire CPS locus of nine additional serotypes (serotypes 3, 4, 5, 8, 10, 16, 19, 23, and 25) and the partial CPS locus of 15 additional serotypes (serotypes 6, 11, 12, 13, 15, 17, 18, 20, 21, 22, 24, 27, 28, 29, 30, and 31) were sequenced [37,51,, 44, 50–52]. Okura et al. [19] completed the sequences of all 33 serotypes, allowing for the development of several multiplex PCR assays for identifying all 33 S. suis serotypes [43, 53, 54]. This method is based on the wzy gene, which is the only serotype-specific gene contained in all serotypes [43]. Real-time PCR [55] and luminex xTAG [56] assays have also been used for the identification of all 33 serotypes. Although all the known serotypes of S. suis can be rapidly and simply identified by molecular biology methods, serotypes 2 and 1/2, as well as serotypes 1 and 14, cannot be differentiated by these methods due to the similar gene content of their respective CPS loci. Athey et al. [57] attempted to differentiate these serotypes from short-read whole-genome sequencing (WGS) data with a bioinformatics pipeline running on a Linux operating system. The short-read WGS data was aligned to the database containing 27 S. suis CPS loci to determine serotype. To differentiate serotype pairs 1 and 14 or 2 and 1/2, the pipeline used custom scripts that identified single-nucleotide polymorphisms in the cpsK gene of every strain identified as a putative serotype 1 or 14 and 2 or 1/2, as well as translation of the amino acid sequences for each strain. The pipeline assigned the final serotype for the strain based on the amino acid present at position 161 of the predicted translated sequence of the cpsK gene. The method is also useful for identifying untypable isolates. Additionally, multilocus sequence typing (MLST), sequence typing (ST), and virulence factor content can also be determined using short-read WGS data. Recent advances in WGS technologies permit the rapid and cost-effective sequencing of hundreds of bacterial genomes. This WGS data alignment method is extremely useful for serotyping S. suis isolates.
Discovery of novel serotypes
In recent years, an increasing number of S. suis isolates have been found that are non-serotypable by serological and/or PCR methods; about 283 nonserotypable isolates have been reported so far (Table S1). Most of the strains were isolated from China. Except for 141 isolates without seroagglutination test results, 42.25% (60/142) of isolates were multi-agglutinating, 28.87% (41/142) of isolates were acapsular, and 17.61% (25/142) of isolates were auto-agglutinating. The other 16 isolates were negative with all 33 serotypes of anti-serum in agglutinating reactions. Most of the 283 non-serotypable isolates (74.20%) were also negative for all the known serotypes using PCR methods. This shows that there are more undiscovered serotypes of S. suis in nature. To identify novel serotypes, agglutination and reversed agglutination (i.e., reaction of the anti-serum to the isolates and the strains with known serotypes) tests should be performed [33]. Additionally, the CPS locus should be sequenced and compared with CPS loci of known serotypes. Of the 283 non-serotypable isolates, only one isolate (CZ130302) was identified as a novel serotype (Chz) using complete identification tests [18••]. The other 97 isolates that have had their CPS loci sequenced were only identified by the agglutination tests and not in the reverse agglutination tests. We predict that the number of S. suis serotypes will increase in the near future.
Conclusions
The relationship between capsule antigenicity and CPS locus genes is very complex. The capsule structure and antigenicity are different for the two serotype pairs (i.e., 1 and 14, 2 and 1/2) with similar CPS loci sequences [9•]. Mutations in the CPS locus, even a single base pair, can change the capsular phenotype [58]. In serotype 1, the gene corresponding to a glycosyltransferase gene (cps14G) in serotype 14 appears to be disrupted by a single base deletion and a resulting frameshift mutation. Such a point mutation could contribute to the antigenic differences between serotypes 1 and 14 [19]. In addition to those in the CPS locus, other known or unknown genes could regulate capsular phenotype [59]. Three serologically un-typeable isolates (i.e., HuN6, HN144, and SS39) were positive for serotype 5 PCR typing (Table S1). Compared to the serotype 5 reference strains, the CPS loci of the three serologically un-typeable isolates contain a longer wcdA gene, which is associated with the translation of a Cap-Dlike protein lengthened at the C terminus. The length difference of the CapD-like protein may change protein activation and influence the anchoring of the capsule to the cell wall envelope, leading to acapsular, multi-agglutinating, or auto-agglutinating reactions [44]. In 2013, 16 S. suis serotype 21/29 strains from healthy pigs were reported [43]. The serotype 21/29 strains that were positive for serotype 29 PCR could agglutinate with anti-sera for both serotypes 29 and 21. The CPS genes of serotype 21/29 strains are highly similar to those of serotype 29, with the exception of cpsH and cpsI. cpsH and cpsI of serotype 21/29 strains are highly similar to serotype 21 strains; meanwhile, the cpsH and cpsI of serotype 21 strains shared no similarities with serotype 29 strains. Thus, PCR results can differ from those of agglutination tests. According to the original rule of S. suis serotyping, positive PCR results cannot solely determine if the isolate belongs to an identical serotype. Although the PCR method is simple, rapid, and sensitive, it cannot be the only method for identifying S. suis serotypes; serological methods should be used to verify the PCR results. WGS and CPS locus sequencing are also powerful tools for identifying known and unknown serotypes, as well as understanding potential mechanisms driving capsular variation. Based on the discoveries of novel serotypes and increase knowledge on capsules and CPS loci of known and novel serotypes, serotyping methods should be improved for better serotype identification of S. suis.
References
Recently published papers of particular interest have been highlighted as: • Of importance •• Of major importance
Gottschalk M, Xu J, Calzas C, Segura M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010;5(3):371–91. doi:10.2217/fmb.10.2.
Outbreak associated with Streptococcus suis in pigs, China. Wkly Epidemiol Rec. 2005;80(32):269–70.
Wertheim HF, Nghia HD, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48(5):617–25. doi:10.1086/596763.
Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21(6):381–407.
Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol. 2005;107(1–2):63–9. doi:10.1016/j.vetmic.2005.01.003.
Vinogradov E, Goyette-Desjardins G, Okura M, Takamatsu D, Gottschalk M, Segura M. Structure determination of Streptococcus suis serotype 9 capsular polysaccharide and assignment of functions of the cps locus genes involved in its biosynthesis. Carbohydr Res. 2016;433:25–30. doi:10.1016/j.carres.2016.07.005.
Van Calsteren MR, Gagnon F, Calzas C, Goyette-Desjardins G, Okura M, Takamatsu D, et al. Structure determination of Streptococcus suis serotype 14 capsular polysaccharide. Biochem Cell Biol. 2013;91(2):49–58. doi:10.1139/bcb-2012-0036.
Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem Cell Biol. 2010;88(3):513–25. doi:10.1139/o09-170.
• Van Calsteren MR, Goyette-Desjardins G, Gagnon F, Okura M, Takamatsu D, Roy R, et al. Explaining the Serological Characteristics of Streptococcus suis Serotypes 1 and 1/2 from Their Capsular Polysaccharide Structure and Biosynthesis. J Biol Chem. 2016;291(16):8387–98. doi:10.1074/jbc.M115.700716. The paper revealed the reason to the two-way cross-reaction of serotype 1 and 2.
Reams RY, Glickman LT, Harrington DD, Thacker HL, Bowersock TL. Streptococcus suis infection in swine: a retrospective study of 256 cases. Part II. Clinical signs, gross and microscopic lesions, and coexisting microorganisms. J Vet Diagn Invest. 1994;6(3):326–34.
Wei Z, Li R, Zhang A, He H, Hua Y, Xia J, et al. Characterization of Streptococcus suis isolates from the diseased pigs in China between 2003 and 2007. Vet Microbiol. 2009;137(1-2):196–201. doi:10.1016/j.vetmic.2008.12.015.
Messier S, Lacouture S, Gottschalk M. Distribution of Streptococcus suis capsular types from 2001 to 2007. Can Vet J. 2008;49(5):461–2.
Aarestrup FM, Jorsal SE, Jensen NE. Serological characterization and antimicrobial susceptibility of Streptococcus suis isolates from diagnostic samples in Denmark during 1995 and 1996. Vet Microbiol. 1998;60(1):59–66.
Wu Z, Zhang W, Lu C. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb Pathog. 2008;45(3):159–66. doi:10.1016/j.micpath.2008.04.009.
Beineke A, Bennecke K, Neis C, Schroder C, Waldmann KH, Baumgartner W, et al. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet Microbiol. 2008;128(3–4):423–30.
Nomoto R, Maruyama F, Ishida S, Tohya M, Sekizaki T, Osawa R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. nov. Int J Syst Evol Microbiol. 2015;65(Pt 2):438–43. doi:10.1099/ijs.0.067116-0.
le Tien HT, Nishibori T, Nishitani Y, Nomoto R, Osawa R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet Microbiol. 2013;162(2–4):842–9. doi:10.1016/j.vetmic.2012.11.001.
•• Pan Z, Ma J, Dong W, Song W, Wang K, Lu C, et al. Novel Variant Serotype of Streptococcus suis Isolated from Piglets with Meningitis. Appl Environ Microbiol. 2015;81(3):976–85. doi:10.1128/AEM.02962-14. A novel serotype of S.suis was identified from clinical sample.
Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol. 2013;79(8):2796–806. doi:10.1128/AEM.03742-12.
Lancefield RC. A Serological Differentiation of Human and Other Groups of Hemolytic Streptococci. J Exp Med. 1933;57(4):571–95.
Demoor CE. Septicaemic Infections in Pigs, Caused by Haemolytic Streptococci of New Lancefield Groups Designated R, S, and T. Antonie Van Leeuwenhoek. 1963;29:272–80.
Elliott SD. Streptococcal infection in young pigs. I. An immunochemical study of the causative agent (PM streptococcus). J Hyg (Lond). 1966;64(2):205–12.
Windsor RS, Elliott SD. Streptococcal infection in young pigs. IV. An outbreak of streptococcal meningitis in weaned pigs. J Hyg. 1975;75(1):69–78.
Perch B, Pedersen KB, Henrichsen J. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol. 1983;17(6):993–6.
Kilpper-Balz R, Schleifer KH. Streptococcus suis sp. nov. nom. rev. Int J Syst Bacteriol. 1987;37(2):160–2.
Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27(12):2633–6.
Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29(11):2590–4.
Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. Description of six new capsular types (29–34) of Streptococcus suis. J Vet Diagn Invest. 1995;7(3):405–6.
•• Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, et al. Current Taxonomical Situation of Streptococcus suis. Pathogens. 2016;5(3):45. doi:10.3390/pathogens5030045.
Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese LA, et al. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48(Pt 2):581–9.
Brousseau R, Hill JE, Prefontaine G, Goh SH, Harel J, Hemmingsen SM. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl Environ Microbiol. 2001;67(10):4828–33.
Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi:10.1146/annurev.micro.50.1.285.
Higgins R, Gottschalk M. An update on Streptococcus suis identification. J Vet Diagn Invest. 1990;2(3):249–52.
Perch B, Kjems E, Slot P, Pedersen KB. Biochemical and serological properties of R, S, and RS streptococci. Acta Pathol Microbiol Scand B. 1981;89(3):167–71.
Charland N, Kellens JT, Caya F, Gottschalk M. Agglutination of Streptococcus suis by sialic acid-binding lectins. J Clin Microbiol. 1995;33(8):2220–1.
Smith HE, Veenbergen V, van der Velde J, Damman M, Wisselink HJ, Smits MA. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J Clin Microbiol. 1999;37(10):3146–52.
Wang K, Fan W, Cai L, Huang B, Lu C. Genetic analysis of the capsular polysaccharide synthesis locus in 15 Streptococcus suis serotypes. FEMS Microbiol Lett. 2011;324(2):117–24. doi:10.1111/j.1574-6968.2011.02394.x.
Wessels MR. Biology of streptococcal capsular polysaccharides. Soc Appl Bacteriol Symp Ser. 1997;26:20S–31.
Segura M, Zheng H, de Greeff A, Gao GF, Grenier D, Jiang Y, et al. Latest developments on Streptococcus suis: an emerging zoonotic pathogen: part 1. Future Microbiol. 2014;9(4):441–4. doi:10.2217/fmb.14.14.
Segura M, Zheng H, de Greeff A, Gao GF, Grenier D, Jiang Y, et al. Latest developments on Streptococcus suis: an emerging zoonotic pathogen: part 2. Future Microbiol. 2014;9(5):587–91. doi:10.2217/fmb.14.15.
Wang K, Zhang W, Li X, Lu C, Chen J, Fan W, et al. Characterization of Streptococcus suis isolates from slaughter swine. Curr Microbiol. 2013;66(4):344–9. doi:10.1007/s00284-012-0275-4.
Zheng H, Ji S, Liu Z, Lan R, Huang Y, Bai X, et al. Eight novel capsular polysaccharide synthesis gene loci identified in nontypeable streptococcus suis isolates. Appl Environ Microbiol. 2015;81(12):4111–9. doi:10.1128/AEM.00315-15.
Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, et al. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One. 2013;8(8), e72070. doi:10.1371/journal.pone.0072070.
Wang K, Sun X, Lu C. Development of rapid serotype-specific PCR assays for eight serotypes of Streptococcus suis. J Clin Microbiol. 2012;50(10):3329–34. doi:10.1128/JCM.01584-12.
Zheng H, Ji S, Lan R, Liu Z, Bai X, Zhang W, et al. Population analysis of Streptococcus suis isolates from slaughtered swine by use of minimum core genome sequence typing. J Clin Microbiol. 2014;52(10):3568–72. doi:10.1128/JCM.00536-14.
Smith HE, van Bruijnsvoort L, Buijs H, Wisselink HJ, Smits MA. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol Lett. 1999;178(2):265–70.
Wisselink HJ, Joosten JJ, Smith HE. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J Clin Microbiol. 2002;40(8):2922–9.
Marois C, Bougeard S, Gottschalk M, Kobisch M. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J Clin Microbiol. 2004;42(7):3169–75.
Silva LM, Baums CG, Rehm T, Wisselink HJ, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115(1–3):117–27.
Wang K, Fan W, Wisselink H, Lu C. The cps locus of Streptococcus suis serotype 16: development of a serotype-specific PCR assay. Vet Microbiol. 2011;153(3–4):403–6. doi:10.1016/j.vetmic.2011.05.050.
Wang K, Lu C, Fan W. Cloning and analysis of the conserved regions in the capsular polysaccharides synthesis locus of Streptococcus suis. Wei Sheng Wu Xue Bao. 2012;52(5):573–80.
Kerdsin A, Dejsirilert S, Akeda Y, Sekizaki T, Hamada S, Gottschalk M, et al. Fifteen Streptococcus suis serotypes identified by multiplex PCR. J Med Microbiol. 2012;61(Pt 12):1669–72. doi:10.1099/jmm.0.048587-0.
Okura M, Lachance C, Osaki M, Sekizaki T, Maruyama F, Nozawa T, et al. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol. 2014;52(5):1714–9. doi:10.1128/JCM.03411-13.
Kerdsin A, Akeda Y, Hatrongjit R, Detchawna U, Sekizaki T, Hamada S, et al. Streptococcus suis serotyping by a new multiplex PCR. J Med Microbiol. 2014;63(Pt 6):824–30. doi:10.1099/jmm.0.069757-0.
Srinivasan V, McGee L, Njanpop-Lafourcade BM, Moisi J, Beall B. Species-specific real-time PCR assay for the detection of Streptococcus suis from clinical specimens. Diagn Microbiol Infect Dis. 2016;85(2):131–2. doi:10.1016/j.diagmicrobio.2016.02.013.
Bai X, Liu Z, Ji S, Gottschalk M, Zheng H, Xu J. Simultaneous detection of 33 Streptococcus suis serotypes using the luminex xTAG(R) assay. J Microbiol Methods. 2015;117:95–9. doi:10.1016/j.mimet.2015.07.018.
Athey TB, Teatero S, Lacouture S, Takamatsu D, Gottschalk M, Fittipaldi N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016;16(1):162. doi:10.1186/s12866-016-0782-8.
Melchiorre S, Camilli R, Pietrantoni A, Moschioni M, Berti F, Del Grosso M, et al. Point mutations in wchA are responsible for the non-typability of two invasive Streptococcus pneumoniae isolates. Microbiology. 2012;158(Pt 2):338–44. doi:10.1099/mic.0.054270-0.
Fittipaldi N, Harel J, D'Amours B, Lacouture S, Kobisch M, Gottschalk M. Potential use of an unencapsulated and aromatic amino acid-auxotrophic Streptococcus suis mutant as a live attenuated vaccine in swine. Vaccine. 2007;25(18):3524–35.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Kaicheng Wang, Zongfu Wu, Huochun Yao, Yuan Qiu, and Chengping Lu declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Bacteriology
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM S1
(DOC 624 kb)
Rights and permissions
About this article
Cite this article
Wang, K., Wu, Z., Yao, H. et al. Identification and Detection of Serotype-Specific Genes: Effective Serotyping of Streptococcus suis . Curr Clin Micro Rpt 4, 29–35 (2017). https://doi.org/10.1007/s40588-017-0055-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40588-017-0055-9