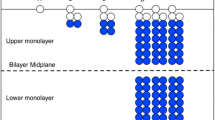
Abstract
We report on the results of particle-based, coarse-grained molecular dynamics simulations of amphiphilic lipid molecules in aqueous environment where the membrane structures at equilibrium are subsequently exposed to strong shock waves, and their damage is analyzed. The lipid molecules self-assemble from unbiased random initial configurations to form stable bilayer membranes, including closed vesicles. During self-assembly of lipid molecules, we observe several stages of clustering, starting with many small clusters of lipids, gradually merging together to finally form one single bilayer membrane. We find that the clustering of lipids sensitively depends on the hydrophobic interaction \(h_\mathrm{c}\) of the lipid tails in our model and on temperature T of the system. The self-assembled bilayer membranes are quantitatively analyzed at equilibrium with respect to their degree of order and their local structure. We also show that—by analyzing the membrane fluctuations and using a linearized theory— we obtain area compression moduli \(K_\mathrm{A}\) and bending stiffnesses \(\kappa _\mathrm{B}\) for our bilayer membranes which are within the experimental range of in vivo and in vitro measurements of biological membranes. We also discuss the density profile and the pair correlation function of our model membranes at equilibrium which has not been done in previous studies of particle-based membrane models. Furthermore, we present a detailed phase diagram of our lipid model that exhibits a sol–gel transition between quasi-solid and fluid domains, and domains where no self-assembly of lipids occurs. In addition, we present in the phase diagram the conditions for temperature T and hydrophobicity \(h_\mathrm{c}\) of the lipid tails of our model to form closed vesicles. The stable bilayer membranes obtained at equilibrium are then subjected to strong shock waves in a shock tube setup, and we investigate the damage in the membranes due to their interaction with shock waves. Here, we find a transition from self-repairing membranes (reducing their damage after impact) and permanent (irreversible) damage, depending on the shock front speed. The here presented idea of using coarse-grained (CG) particle models for soft matter systems in combination with the investigation of shock-wave effects in these systems is a quite new approach.













Similar content being viewed by others
References
McNeil PL, Terasaki M (2001) Coping with the inevitable: how cells repair a torn surface membrane. Nat Cell Biol 3(5):E124–E129
Schmidt M, Kahlert U, Wessolleck J, Maciaczyk D, Merkt B, Maciaczyk J, Osterholz J, Nikkhah G, Steinhauser MO (2014) Characterization of s setup to test the impact of high-amplitude pressure waves on living cells. Nat Sci Rep 4:3849-1–3849-9
Gambihler S, Delius M, Ellwart JW (1992) Transient increase in membrane permeability of L1210 cells upon exposure to lithotripter shock waves in vitro. Die Naturwissenschaften 79:328–329
Gambihler S, Delius M, Ellwart JW (1994) Permeabilization of the plasma membrane of L1210 mouse leukemia cells using lithotripter shock waves. J Membr Biol 141:267–275
Kodama T, Doukas AG, Hamblin MR (2002) Shock wave-mediated molecular delivery into cells. Biochim Biophys Acta 1542:186–194
Bao G, Suresh S (2003) Cell and molecular mechanics of biological materials. Nat Mater 2:715–725
Tieleman DP, Leontiadou H, Mark AE, Marrink S-J (2003) Simulation of pore formation in lipid bilayers by mechanical stress and electric fields. J Am Chem Soc 125:6382–6383
Sundaram J, Mellein BR, Mitragotri S (2003) An experimental and theoretical analysis of ultrasound-induced permeabilization of cell membranes. Biophys J 84:3087–3101
Doukas AG, Kollias N (2004) Transdermal drug delivery with a pressure wave. Adv Drug Deliv Rev 56:559–579
Coussios C-C, Roy RA (2008) Applications of acoustics and cavitation to noninvasive therapy and drug delivery. Ann Rev Fluid Mech 40:395–420
Prausnitz MR, Langer R (2008) Transdermal drug delivery. Nat Biotechnol 26:1261–1268
Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker CJ (2011) The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater 10:389–397
Koshiyama K, Wada S (2011) Molecular dynamics simulations of pore formation dynamics during the rupture process of a phospholipid bilayer caused by high-speed equibiaxial stretching. J Biomech 44:2053–2058
Phillips R, Kondev J, Theriot J (2009) Physical biology of the cell. Garland Science. Taylor and Francis Group, New York
Alberts B, Bray D, Hopkin K, Jonson A, Lewis J, Raff M, Roberts K, Walter P (2010) Essential cell biology. Garland Science. Taylor and Francis Group, New York
Alberts B, Bray D, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P (2000) Molecular biology of the cell. Garland Science. Taylor and Francis Group, New York
Steinhauser MO (2006) Computational methods in polymer physics. Recent Res Dev Phys 7:59–97
Steinhauser MO (2008) Computational multiscale modeling of fluids and solids—theory and applications. Springer, Berlin
Steinhauser MO, Schneider J, Blumen A (2009) Simulating dynamic crossover behavior of semiflexible linear polymers in solution and in the melt. J Chem Phys 130:164902-1–164902-8
De Weer P (2000) A century of thinking about cell membranes. Annu Rev Physiol 62:919–926
Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175:720–731
Yang K, Ma YQ (2010) Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat Nanotechnol 5:579–583
Das C, Sheikh KH, Olmsted PD, Connell SD (2010) Nanoscale mechanical probing of supported lipid bilayers with atomic force microscopy. Phys Rev E 82:041920-1–041920-6
Edidin M (2003) Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol 4:414–418
Bowick MJ, Travesset A (2001) The statistical mechanics of membranes. Phys Rep 344:255–308
Nelson D, Piran T, Weinberg S (2004) Statistical mechanics of membranes and surfaces. World Scientific Publishing, Singapore
Allen MP, Tildesley DJ (1987) Computer simulation of liquids. Clarendon Press, Oxford
Rapaport DC (2004) The art of molecular dynamics simulation. Cambridge University Press, Cambridge
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824
Steinhauser MO (2013) Computer simulation in physics and engineering. Walter de Gruyter, Berlin
Drouffe JM, Maggs AC, Leibler S (1998) Computer simulations of self-assembled membranes. Science 254:1353–1356
Goetz R, Lipowsky R (1998) Computer simulations of bilayer membranes: self-assembly and interfacial tension. J Chem Phys 108:7397–7409
Noguchi H, Takasu M (2001) Self-assembly of amphiphiles into vesicles: a Brownian dynamics simulation. Phys Rev E 64:041913-1–041913-7
Bourov GK, Bhattacharya A (2005) Brownian dynamics simulation study of self-assembly of amphiphiles with large hydrophilic heads. J Chem Phys 122:44702-1–44702-6
Noguchi H, Gompper G (2006) Dynamics of vesicle self-assembly and dissolution. J Chem Phys 125:164908-1–164908-13
Tribet C, Vial F (2008) Flexible macromolecules attached to lipid bilayers: impact on fluidity, curvature, permeability and stability of the membranes. Soft Matter 4:68–81
Yang S, Qu J (2014) Coarse-grained molecular dynamics simulations of the tensile behavior of a thermosetting polymer. Phys Rev E 90:012601-1–012601-8
Brannigan G, Lin LCL, Brown FLH (2006) Implicit solvent simulation models for biomembranes. Eur Biophys J 35:104–124
Dror RO, Jensen MO, Borhani DW, Shaw DE (2010) Perspectives on: molecular dynamics and computational methods: exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. J Gen Physiol 135:555–562
Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput 8:1542–1555
Skjevik AA, Madej BD, Dickson CJ, Teigen K, Walker RC, Gould IR (2015) All-atom lipid bilayer self-assembly with the AMBER and CHARMM lipid force fields. Chem Commun 51:4402–4405
Steinhauser MO (2012) Modeling dynamic failure behavior in granular and biological materials: emerging new applications. IJATEMA 1:15–29
Steinhauser MO, Schmidt M (2014) Destruction of cancer cells by laser-induced shock waves: recent developments in experimental treatments and multiscale computer simulations. Soft Matter 10:4778–4788
Huber PE, Jenne J, Debus J, Wannenmacher MF, Pfisterer P (1999) A comparison of shock wave and sinusoidal-focused ultrasound-induced localized transfection of HeLa cells. Ultrasound Med Biol 25:1451–1457
Dubinsky TJ, Cuevas C, Dighe MK, Kolokythas O, Hwang JH (2008) High-intensity focused ultrasound: current potential and oncologic applications. Am J Roentgenol 190:191–199
Gevaux D (2010) Physics and the cell. Nat Phys 6:725
Fritsch A, Höckel M, Kiessling T, Nnetu KD, Wetzel F, Zink M, Käs JA (2010) Are biomechanical changes necessary for tumour progression? Nat Phys 6:730–732
Koshiyama K, Kodama T, Yano T, Fujikawa SS (2006) Structural change in lipid bilayers and water penetration induced by shock waves: molecular dynamics simulations. Biophys J 91:2198–2205
Koshiyama K, Kodama T, Yano T, Fujikawa SS (2008) Molecular dynamics simulation of structural changes of lipid bilayers induced by shock waves: effects of incident angles. Biochim Biophys Acta 1778:1423–1428
Lechuga J, Drikakis D, Pal S (2008) Molecular dynamics study of the interaction of a shock wave with a biological membrane. Int J Numer Mech Fluids 57:677–692
Sridhar A, Srikanth B, Kumar A, Dasmahapatra AK (2015) Coarse-grain molecular dynamics study of fullerene transport across a cell membrane. J Chem Phys 143:024907-1–024907-9
Hofmann A, Ritz U, Rompe JD, Tresch A, Rommens PM (2015) The effect of shock wave therapy on gene expression in human osteoblasts isolated from hypertrophic fracture non-unions. Shock Waves 25:91–102
Hoon Ha C, Cheol Lee S, Kim S, Chung J, Bae H, Kwon K (2015) Novel mechanism of gene transfection by low-energy shock wave. Nat Sci Rep 5:12843-1–12843-13
Zhang Z, Lu L, Noid WG, Krishna V, Pfaendtner J, Voth GA (2008) A systematic methodology for defining coarse-grained sites in large biomolecules. Biophys J 95:5073–5083
Noguchi H (2009) Membrane simulation models from nanometer to micrometer scale. J Phys Soc Jpn 78:041007
Murtola T, Karttunen M, Vattulainen I (2009) Systematic coarse graining from structure using internal states: application to phospholipid/cholesterol bilayer. J Chem Phys 131:055101-1–055101-15
Müller M (2011) Studying amphiphilic self-assembly with soft coarse-grained models. J Stat Phys 145:967–1016
Lyubartsev AP, Rabinovich AL (2010) Recent development in computer simulations of lipid bilayers. Soft Matter 7:25–39
Huang MJ, Kapral R, Mikhailov AS, Chen HY (2012) Coarse-grain model for lipid bilayer self-assembly and dynamics: multiparticle collision description of the solvent. J Chem Phys 137:055101-1–055101-10
May A, Pool R, van Dijk E, Bijlard J, Abeln S, Heringa J, Feenstra KA (2014) Coarse-grained versus atomistic simulations: realistic interaction free energies for real proteins. Bioinformatics 30:326–334
Yuan H, Huang C, Li J, Lykotrafitis G, Zhang S (2010) One-particle-thick, solvent-free, coarse-grained model for biological and biomimetic fluid membranes. Phys Rev E 82:011905-1–011905-8
Noguchi H (2011) Solvent-free coarse-grained lipid model for large-scale simulations. J Chem Phys 134:055101-1–055101-12
Warshel A, Levitt M (1976) Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol 103:227–249
Dünweg B, Reith D, Steinhauser MO, Kremer K (2002) Corrections to scaling in the hydrodynamic properties of dilute polymer solutions. J Chem Phys 117:914–924
Steinhauser MO (2005) A molecular dynamics study on universal properties of polymer chains in different solvent qualities. Part I. A review of linear chain properties. J Chem Phys 122:094901-1–094901-13
Stevens MJ (2004) Coarse-grained simulations of lipid bilayers. J Chem Phys 2004(121):11942–11948
Steinhauser MO, Grass K, Strassburger E, Blumen A (2009) Impact failure of granular materials—non-equilibrium multiscale simulations and high-speed experiments. Int J Plast 25:161–182
Saunders MG, Voth GA (2012) Coarse-graining methods for computational biology. Annu Rev Biophys 42:73–93
Nielsen SO, Lopez CF, Srinivas G, Klein ML (2004) Topical review: coarse grain models and the computer simulation of soft materials. J Phys-Condens Matter 16:481–R512
Pogodin S, Baulin VA (2010) Coarse-grained models of phospholipid membranes within the single chain mean field theory. Soft Matter 6:2216–2226
Wang Y, Sigurdsson JK, Brandt E, Atzberger PJ (2013) Dynamic implicit-solvent coarse-grained models of lipid bilayer membranes: fluctuating hydrodynamics thermostat. Phys Rev E 88:023301-1–023301-5
Farago O (2003) ”Water-free” computer model for fluid bilayer membranes. J Chem Phys 119:596–605
Brannigan G, Philips PF, Brown FLH (2005) Flexible lipid bilayers in implicit solvent. Phys Rev E 72:011915-1–011915-4
Sodt AJ, Head-Gordon T (2010) An implicit solvent coarse-grained lipid model with correct stress profile. J Chem Phys 132:205103-1–205103-8
Shih AY, Arkhipov A, Freddolino PL, Schulten K (2006) Coarse grained protein-lipid model with application to lipoprotein particles. J Phys Chem B 110:3674–3684
Noguchi H, Takasu M (2002) Adhesion of nanoparticles to vesicles: a brownian dynamics simulation. Biophys J 83:299–308
Cooke IR, Deserno M (2005) Solvent-free model for self-assembling fluid bilayer membranes: stabilization of the fluid phase based on broad attractive tail potentials. J Chem Phys 123:224710-1–224710-13
Izvekov S, Voth GA (2009) Solvent-free lipid bilayer model using multiscale coarse-graining. J Phys Chem B 113:4443–4455
Wang ZJ, Frenkel D (2005) Modeling flexible amphiphilic bilayers: a solvent-free off-lattice Monte Carlo study. J Chem Phys 122:234711-1–234711-8
Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML (2001) A coarse grain model for phospholipid simulations. J Phys Chem B 105:4464–4470
Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Moore PB, Klein ML (2001) Simulations of phospholipids using a coarse grain model. J Phys Chem B 105:9785–9792
Marrink SJ, de Vries AH, Mark AE (2003) Coarse grained model for semiquantitative lipid simulations. J Phys Chem B 108:750–760
Brannigan G, Brown FLH (2004) Solvent-free simulations of fluid membrane bilayers. J Chem Phys 120:1059–1071
Cooke IR, Kremer K, Deserno M (2005) Tunable generic model for fluid bilayer membranes. Phys Rev E 72:011506-1–011506-4
Ganzenmüller GC, Hiermaier S, Steinhauser MO (2012) Energy-based coupling of smooth particle hydrodynamics and molecular dynamics with thermal fluctuations. Eur Phys J Spec Top 206:51–60
Steinhauser MO (2008) Static and dynamic scaling of semiflexible polymer chains—a molecular dynamics simulation study of single chains and melts. Mech Time Depend Mater 12:291–312
Griffiths G (2007) Cell evolution and the problem of membrane topology. Nat Rev Mol Cell Biol 8:1018–1024
Yang K, Ma YQ (2012) Computer simulations of fusion, fission and shape deformation in lipid membranes. Soft Matter 8:606–618
Chernomordirk L, Kozlov MM, Zimmerberg J (1995) Lipids in biological membrane-fusion. J Membr Biol 146:1–14
Helfrich W (1973) Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C 28:693–703
Seifert U (1997) Configurations of fluid membranes and vesicles. Adv Phys 46:13–137
Goetz R, Gompper G, Lipowsky R (1999) Mobility and elasticity of self-assembled membranes. Phys Rev Lett 82:221–224
Olbrich K, Rawicz W, Needham D, Evans E (2000) Water permeability and mechanical strength of polyunsaturated lipid bilayers. Biophys 79:321–327
Lee CH, Lin WC, Wang J (2001) All-optical measurements of the bending rigidity of lipid-vesicle membranes across structural phase transitions. Phys Rev E 64:020901-1–02091-4
Doukas AG, McAuliffe DJ, Lee S, Venugopalan V, Flotte TJ (1995) Physical factors involved in stress-wave-induced cell injury: the effect of stress gradient. Ultrasound Med Biol 21:961–967
Kodama T, Hamblin MR, Doukas AG (2000) Cytoplasmic molecular delivery with shock waves: importance of impulse. Biophys J 79:1821–1832
Hansen JP, McDonald IR (2006) Theory of simple liquids. Academic Press, Oxford
Ganzenmüller GC, Hiermaier S, Steinhauser MO (2011) Shock-wave induced damage in lipid bilayers: a dissipative particle dynamics simulation study. 7:4307–4317
Funding
This study was funded by the Fraunhofer-Gesellschaft, e.V., Germany, under grant no. 400017 “Extracorporeal, Focused Ultrasound Therapy: Effectiveness, Simulation, and Planning of New Therapies” and grant no. 600016, Vintage Class Program: “Shock Wave Induced Destruction of Tumor Cells”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (mp4 22464 KB)
Rights and permissions
About this article
Cite this article
Steinhauser, M.O., Schindler, T. Particle-based simulations of bilayer membranes: self-assembly, structural analysis, and shock-wave damage. Comp. Part. Mech. 4, 69–86 (2017). https://doi.org/10.1007/s40571-016-0126-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40571-016-0126-3