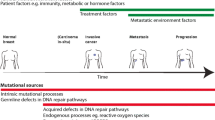
Abstract
Cancer is the most challenging disease of our time with increasing numbers of new cases each year, worldwide. Great achievements have been reached in cancer research through deep sequencing which helped define druggable targets. However, the still-evolving targeted therapy suffers resistance suggesting that DNA mutations considered as drivers may not have a role in tumor initiation. The present work discusses the role of DNA mutations as drivers and passengers in cancer initiation and development. First, it is important to discern the role of these DNA mutations as initiating events causing cancer or as contributors crucial for the development of a tumor once it has initiated. Second, breast cancer shown here illustrates how identification of DNA mutations in cancerous cells has influenced our approach for anti-cancer drug design. The cancer trilogy we have reached and described as: initial drug; resistance/recurrence; drug/treatment combinations, calls for a paradigm shift. To design more effective cancer drugs with durable and positive outcome, future cancer research needs to move beyond the sequencing era and explore changes which are taking place in cancer cells at levels other than the DNA. Evolutionary constraints may be acting as a barrier to preserve the human species from being transformed and, for that matter, all multi-cellular species which can incur cancer. Furthermore, mutations in the DNA do occur and for a multitude of reasons but without necessarily causing cancer. New directions will draw themselves when more focus is given to the event responsible for the switch of a cell from normalcy to malignancy. Until then, targeted therapy will certainly continue to improve the outcome of patients; however, it is unlikely to eradicate breast cancer depicted here.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is the most challenging disease of our time, with increasing numbers of new cases each year worldwide. The number of drugs designed to treat cancer is also increasing [1], but without achieving the intended goal of curing cancer. Moreover, accumulated evidence shows that cancer therapy today has come to what looks like an endless battle as new drugs seem only to lead to new paths of resistance. Resistance mechanisms so far identified include, among others, drug inactivation, drug target alteration, drug efflux, DNA damage repair, cell death inhibition and epithelial–mesenchymal transition [2]. The obvious question here is whether we are battling cancer from the right front.
No doubt, cancer is a complicated disease and, no doubt, enormous progress has been made in cancer research that has shed light on almost every biological aspect of this disease. The cancer genome sequencing project allowed the discovery of oncogenic mutations and gave valuable insights into the genomics of cancer. Today, the understanding of the cancer genome has become an important aspect of the clinicians’ decision to select the best treatment available for each cancer patient. Moreover, the establishment of the Cancer Genome Atlas (TCGA) [3] is the most important scientific advance of the century and analysis of cancer genomes continues to supply valuable information with clinical and therapeutic implications. Cataloging genetic mutations responsible for cancer, TCGA project, which begun in 2005, used several techniques to provide a large, statistically significant data set for further discovery. Today, whether searching to understand the pathogenesis of tumor development such as in lung squamous cell carcinoma [4] or in oropharyngeal carcinomas [5], consulting TGCA for that end is of paramount importance.
All this progress is still, however, not enough because we are not yet done with cancer. In the midst of the struggle to cure cancer and in the midst of the repeated defeat registered in cancer therapy, one asks whether going after DNA mutations can one day lead us to inhibit the rising of cancerous cells. Shrinking a tumor is one thing, but preventing the rising of transformed cancerous cells which form tumors is another thing totally different. Shrinking a tumor translates into a symptom-oriented treatment while preventing the rise of cancerous cells translates into a cure of cancer.
This original work discusses the role of DNA mutations as drivers and passengers in cancer initiation and development. It is deemed important to discern the role of these DNA mutations and whether they are the cause that initiates cancer or arise as a consequence of tumor formation and contribute to the growth and the development of the tumor once it had initiated. It also discusses through an example of breast cancer, how the present understanding of driver/passenger mutations has impacted our strategy for cancer drug development and cancer therapy.
This article is based on previously conducted studies and does not involve any new studies of human or animal subjects performed by any of the authors.
Tumor Genome Sequencing: a Giant Step in Cancer Research
The sequencing of the entire human genome completed in 2003 opened the doors to explore cancer genomes. As technology made more progress with large-scale genome sequencing techniques, the Cancer Genome Anatomy Project (CGAP) set new goals to determine gene expression profiles of cancerous, precancerous and normal tissues [6]. The objective behind sequencing cancer genomes is to look for driver mutations, which increase the mutation rate in the cell, leading to a more rapid evolution of the tumor and metastases formation [7]. The rationale behind a mutated gene as causal in cancer is based on the observation that the number and pattern of mutations in affected genes were found to be very unlikely the result of chance [8]. Therefore, mutational data obtained through sequencing can be used to identify candidate cancer genes that are most likely to be the drivers. Moreover, it is reasonable to suggest that genes that are mutated more frequently than predicted by chance are more likely the drivers [9].
Some of the identified mutations are called passengers as they do not drive progression to metastatic disease [10], whereas others called genetic drivers [11, 12] are considered crucial for cancer cell survival and growth. Vogelstein et al. defined a driver gene mutation as a mutation that directly or indirectly confers a selective growth advantage to the cell in which it occurs, whereas a passenger mutation is a mutation that has no direct or indirect effect on the selective growth advantage of the cell in which it occurred [13].
Although it is difficult to determine driver mutations from DNA sequences alone, drivers tend to be the most commonly shared mutations between tumors and cluster around known oncogenes and also tend to be non-silent [14], whereas passenger mutations are randomly distributed throughout the genome. Clones bearing driver mutations are assumed to be positively selected in the evolution of neoplasia to invasive and advanced cancer [10].
The sequencing of cancer genomes has thus helped identify specific and unique changes in cancer patients and based on this information, a personalized therapeutic strategy [15, 16] became possible. Additionally, while genome sequencing can provide important information on patients with rare or novel tumor types, translating such information into clinical treatment strategy is often complicated [15, 16]. Moreover, malignancies are known to have a spectrum of gene mutations which can affect several metabolic pathways, complicating the task further. Cancer patients may not be able to afford the costs of personalized therapy while others may lack access to such treatment option.
Nonetheless, it is important to highlight the benefits of the sequencing data and mutation cataloging in clinics. The identification of cancer mutations led the way to targeted therapy which is in its full development, helping clinicians make major decision concerning each patient’s treatment. Through such personalized medicine, many patients’ outcomes have been significantly improved. In lung cancer, the discovery of the anaplastic lymphoma kinase gene (ALK) rearrangements and the subsequent development of crizotinib, an oral tyrosine kinase inhibitor targeting ALK for a selected ALK-positive patient group, is an example of such a successful biomarker-driven drug development [17]. Shaw et al. reported a median progression-free survival of 7.7 months in the crizotinib group compared to 3.0 months in the chemotherapy group. The authors have concluded that crizotinib is superior to standard chemotherapy in patients with previously treated, advanced non-small-cell lung cancer (NSCLC) with ALK rearrangement [17].
The problem, however, resides in the continuous trend of cancer cells’ evolution leading to the appearance of new mutations while on treatment or after treatment leading thus to drug resistance or recurrence as clearly shown in NSCLC targeted with EGFR-TK inhibitors. In selected NSCLC patients, the constitutive activation of EGFR signaling caused by gene mutations/amplification or both is responsible for the oncogenic behavior and poor prognosis of NSCLC [18]. Mutations in the EGFR gene which occur in ~10–35% of NSCLC patients are considered important drivers in NSCLC [19] and have played a pivotal role as rational targets in NSCLC-targeted therapy, revolutionizing lung cancer care and treatment as a consequence. Three generations of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have thus far been developed (reviewed in [20]). However, most patients treated with EGFR-TKIs develop resistance within 9–14 months [21]. Oxnard et al. have reported that acquired resistance mechanisms of AZD9291 in patients with EGFRT790M-mutant NSCLC who failed treatment with first-generation EGFR TKIs include the loss of EGFRT790M-mutant clones plus alternative pathway activation or histologic transformation and EGFR ligand-dependent activation [22].
The question which remains to be answered is how to overcome resistance and recurrence phenomena in targeted and personalized medicine? This resistance barrier is perhaps teaching us to take a major turn in cancer research and redefine the etiology of cancer in order to redefine its treatment.
The Driver–Passenger Point of View
One of the first sequencing studies estimated that individual colorectal cancers contain about 100 non-synonymous mutations and that as many as 20 of the mutated genes in individual cancers might play a causal role in the neoplastic process [23]. Data from this study also showed that the mutational patterns among colorectal cancers from different patients are diverse. In another sequencing study, it has been estimated that the average tumor carries around 80 somatic mutations, fewer than 15 of which are expected to be drivers [9]. However, studies on melanoma have revealed a complex genetic background where melanoma genomes include a high rate of mutation, a complex copy number landscape, UV-related C > T transitions, and frequent genetic alterations in well-known drivers of melanoma genesis such as BRAF, NRAS, TP53, CDKN2A, and PTEN [11, 24,25,26].
More recent studies have shown that an average cancer of the breast or the colon can harbor about 60–70 protein-altering mutations, of which 3 or 4 may be driver mutations while the remaining may be passenger mutations [13], and that at least 125 mutated driver genes have been identified among 3284 sequenced tumor genomes. Another study conducted by Ding et al. on 97 tumors from 96 melanoma patients revealed truncation mutations, in tumor suppressors, protein phosphatases, as well as genes involved in chromatin remodeling, which co-occur with BRAF and NRAS mutations [27].
From the driver–passenger point of view, mutations in the BRAF gene, mainly the V600E mutation, are considered a driver leading to constitutive activation of the MAP kinase pathway and increase in growth signals [28]. However, the BRAF V600E mutation has been identified in only 40–45% of metastatic melanomas [29], which is lower compared with melanocytic nevi [30, 31]. Moreover, whole-genome sequencing of 100 gastric tumors, a heterogeneous cancer with diverse molecular and histological subtypes, revealed more than seven driver genes including known genes such as TP53, ARID1A and CDH1 and newly identified ones (MUC6, CTNNA2, GLI3, RNF43) [32].
These observations clearly show that drivers are many and diverse and the main question here is how to conquer cancer with these many mutations in a single type of cancer? How many drivers can we target at once in a given patient without increasing toxicity? Most importantly, though, is what really needs to be targeted in cancer? The end product, i.e. the tumor, which can be formed through the hijacking of several different metabolic pathways as shown in the diversity of mutated genes, or the force and entity behind the establishment of these DNA mutations? DNA mutations which shape each tumor can go on endlessly and can change according to their microenvironment, including in the presence of chemotherapeutic drugs and radiation. Running after genes that are mutated in cancer cells may mean running in the same direction with cancer cells, but not in the opposite direction, as it should be in order to stop their proliferation. To stop cancer growth, we should go in the opposite direction, come face-to-face and neutralize the entity behind the establishment of these DNA mutations in cancerous cells. Isn’t this capability of easily mutating that is behind resistance phenomena observed in clinics?
On the other hand, it has been suggested that the failure of current cancer therapies may be the result of the used drugs not targeting what really causes cancer [33]. Also, in the hypothetical settings where the cause of cancer is still unknown and remains to be identified [33], what is, then, the role of these DNA mutations in cancer genesis? Do any of these mutations in driver genes cause the switch from normalcy to malignancy? There is no evidence that such is the case and none of the identified driver DNA mutation causes the switch of a cell from normalcy to malignancy. Moreover, these driver mutations are not found in 100% of tumors in any given cancer type while being absent in their normal counterpart cells. There is, however, ample evidence showing that these identified DNA mutations together contribute to the rising of cancer. The prevalent idea is that a large number of mutations, each associated with a small fitness advantage, drive tumor progression [34]. On the other hand, an important finding showed more passengers (hills) than drivers (mountains) shaping cancer landscapes at least in breast and colon cancers, and passenger rates vary considerably from tumor to tumor [9]. Moreover, studies on myelomas suggested that there are multiple genes, each mutated in a small proportion of tumors that can alter the same signal transduction pathway [35, 36]. Based on these observations, the number of potential drivers, spreading to include a diverse range of genes, appears large enough to qualify as the driving cause in one and the same disease. Furthermore, it is not clear how to separate a driver from a passenger somatic mutation [13].
While these DNA mutations may explain how cancer progresses, they do not explain how it starts. The important question to ask is not how cancer progresses but instead how cancer starts. If the focus is on the tumor and how to shrink it then these DNA mutations in driver genes are indeed important to consider for the design of anti-cancer drugs. However, if we seek long-lasting outcomes in cancer treatment, then the focus should shift from the tumor, formed by a mass of heterogeneous cancerous cells, to the level of a single cancerous cell. Putting more emphasis on tumor growth has led us to design drugs aimed at shrinking those tumors while we lost from sight what really causes a normal cell to become cancerous in the first place.
Looking thus from a different angle, these DNA mutations may be interpreted as consequences of transformation but not the cause of it. Once a normal cell has switched to a cancerous cell, symptoms of that transformation are seen in the form of a number of DNA mutations we came to call drivers and passengers. Each major mutation or set of mutations may indicate the path taken by different cancerous cells on their way to invasion and metastases. Following this line of reasoning, there should be no driver or passenger gene mutation per se, as mutations seen in cancer may be the result of that cellular modification catalyzing the switch from normalcy to malignancy that has yet to be identified [33]. Therefore, the observed mutations in cancer cells may occur to fulfill the malignant character and complete the reprogramming [33] process during transformation.
On the other hand, mutations can occur in the human genome without causing cancer. Mutations found in the FGFR3 have been reported in 18–85% of seborrheic keratoses which are benign lesions with no risk of cancer [37]. Another reason to question the idea of driver mutations, i.e. a modification at the DNA level as causal in cancer, becomes evident in the fact that mutations in BRAF and NRAS, originally qualified as drivers, could not fully explain melanoma oncogenesis, as these same mutations have been found at similar rates in benign nevi [38, 39].
In a study conducted by Poynter JN et al., the role of BRAF/NRAS mutations has been investigated in samples from a case control study of melanoma and a series of benign melanocytic nevi. The study showed evidence of the up-regulation of the mitogen-activated protein kinase pathway in a large percentage of melanocytic lesions, but these mutations have been suggested to be not sufficient for malignant transformation. The authors suggested that BRAF mutations contribute to benign melanocytic hyperplasia, but are likely to contribute to invasive melanoma only in conjunction with other mutations [40]. A more recent study in melanoma, however, showed that BRAF and NRAS co-mutations are not mutually exclusive and that the co-existence of BRAF and NRAS driver mutations in the same melanoma cells resulted in heterogeneity of resistance in response to targeted therapy that was not observed in non-targeted therapy [41].
Inherited DNA Mutations and Cancer
If somatic mutations in driver genes do not fully qualify to directly cause cancer, how about germ line mutations? If the cause of cancer was in the DNA, one should expect a direct effect of the inherited driver mutation(s) to cause cancer and this should not be limited to a predisposition to develop cancer. In the light of this work, predisposition in the presence of a germ line mutation means that the ground is fertile to drive transformation but not to initiate it. Therefore, the presence of a germ line mutation alone is unlikely to cause cancer and limits its effect on predisposition to cancer but is unable by itself to switch a cell from normalcy to malignancy. In this line of reasoning, individuals with germ line mutations in DNA repair genes cannot develop cancer unless exposed to UV radiation [42, 43]. Moreover, the effect of inherited mutations should be systematic and not limited to one organ only, such as the skin in the case of melanoma. And why not? Is not the inherited driver mutation present in each and every cell of the body? Or do we assume that each organ has its own specific driver DNA mutation in order to develop its own cancer?
In this view, seeing BRCA1/2 genes more often mutated in breast cancer, and the BRAF gene more often muted in melanoma, may solely reflect differences in metabolic pathways in these specialized organs and tissues’ functions. It has been suggested that tissue specificity of inherited mutations in BRCA1, for example, may result from BRCA1 being evolutionarily recruited to suppress cancer in breast and ovarian tissues, but not for such a role in the non-susceptible tissues [44]. Based on such tissue functional specialization, the yet unidentified cancer-causing entity acts accordingly to hijack the major metabolic pathway in that specific tissue, rerouting it towards transformation. However, this may not be the rule in cancer as cancer cells are thought to not follow universal laws governing cell division [33], but can take advantage of any shortcuts available, adapting to their microenvironment and resulting, as a consequence, in tumor heterogeneity. This heterogeneity may explain why BRCA1/2 and BRAF gene mutations, for example, are not found in 100% of breast cancers and melanomas, respectively.
It is important to note that several genes with germ line mutations that cause cancer predisposition have been reported to show very few, if any, somatic mutations in sporadic cancers of the same type, such as BRCA1 and BRCA2 in breast cancer [45, 46]. It has also been suggested that genes predisposing to cancer, when inherited in mutant forms in the germ line, stimulate tumorigenesis in indirect ways but do not confer an increase in selective growth advantage [13]. Moreover, inherited PIK3CA-activating mutations are implicated in seborrheic keratosis (16%) and epidermal nevi (27%), with mutations identical to those found in certain malignancies [37]. Furthermore, TP53 germ line mutations lead to Li–Fraumeni syndrome that is associated with a high incidence of cancer [47], and synovial tissues from patients with rheumatoid arthritis had alterations in TP53 (17–46%) that are identical to the mutations seen in malignancies [48, 49]. Therefore, none of these inherited mutations turns on tumorigenesis and causes cancer directly, even when the gene is qualified as a driver as opposed to a passenger gene. Moreover, deficiency in DNA repair proteins due to an inherited mutation increases cancer risks, such as mutations in the TP53 gene [50], but germ line mutations in DNA repair genes were found to be the cause of only 1% of cancers [51].
These observations clearly suggest that the causing event in cancer may not lie in the DNA sequence in a form of mutations. Therefore, how can we rely on these heterogeneous and evolving genomic alterations and design cancer drugs without the risk of being counterattacked with resistance responses?
Chromosome Instability and Cancer Initiation
Chromosome instability (CIN) is a hallmark of most solid tumors [52] with aneuploidy being a direct result of CIN, observed in about 90% of all solid tumors [53] and has been suggested to serve as a driver of cellular transformation [54]. Hereditary cancers are often characterized by the presence of mutations in DNA repair genes, such as BRCA1, BRCA2, MSH2 and MYH, which lead to genomic instability. Genomic instability is present in all stages of cancer, from precancerous lesions, even before TP53 mutations are acquired [55], to advanced cancers [56].
An important finding in mouse model experiments failed to show CIN as a driver in transformation. These experiments often needed an additional genetic alteration for tumorigenesis, such as the deletion of the p53, p21 or p19Arf tumor suppressor genes [57, 58]. Moreover, studies conducted by Silk et al. showed that exacerbating the level of CIN in mice model experiments resulted in enhanced cell death and reduced tumor growth [59]. These results could likely be explained in the light of this work by the absence of the cancer-causing entity in these animal models. The most important question to answer in cancer is: What makes genomic alterations, which include all forms of DNA changes, viable in cancerous cells? Unless we assume that what causes cancer bears the properties to first, allow these DNA alterations to occur, and second, to use them for tumor growth, invasion and metastases. In the absence of the cellular modification that causes cancer, these gross DNA alterations, regardless of their extent and form, are unviable. Another strong argument is seen in humans where it is well known that a large proportion of first-trimester spontaneous abortions are caused by chromosomal disorders, and a correlation between chromosome abnormalities and spontaneous abortion has been documented for decades [60]. These spontaneous abortions could be viewed as a mechanism that has evolved to protect human species from being transformed.
The Evolution Point of View
The aim of this work is not to debate theories on cancer origin and evolutionary models; however, it is the aim of this work to try to answer the question of whether evolution, when defined as that universal law through which living and non-living matter evolve toward higher structures of higher complexity and function [33], could allow DNA, the genetic material defining each living species, to bear the cause of cancer. Moreover, if evolution has not selected for cancer but rather did not hamper its appearance [33], it follows from this understanding that ingraining the cause of cancer in the genetic material under the form of DNA mutations would have had devastating consequences not only to our human species but to all multi-cellular organisms which can incur cancer. This important argument points to the causing event in cancer that may not be carried on DNA sequences. On the other hand, DNA is the core and essence of living species and evolution would not let selfish cancer cells to meddle with it. All living species are their DNA; without this identity, they would not exist in their present forms. And though it can be affected, DNA cannot be transformed or bear a transforming mutation which would cause cancer that transforms and destroys organs’ function, disturbs the homeostasis of the organism and leads to death. By being affected as a secondary event resulting from the cellular modification that causes cancer which has yet to be identified, DNA mutational effects become restrained to the tissues only in which cancer grows, making, hence, the consequence local but not global. Local effects destroy tissues’ function and global effects would affect the species as a whole, destroying and/or transforming it. Moreover, once cancer has initiated, its evolution relies largely on DNA mutations which have been shown to play a key role in both the development of tumors and their response to therapy [61]. Constraints exercised by these DNA mutations on cancer evolution are reviewed by Venkatesan et al. [62].
Trunk and Branch Mutations and the Race to Harness Tumor Heterogeneity
Clonal evolution of tumors has been modeled as a tree with trunk and branch mutations describing intra-tumor heterogeneity. The trunk harbors somatic aberrations thought to arise at early stages of tumor formation, whereas branches harbor somatic aberrations thought to appear later at separation of branches [63, 64]. Trunk or clonal mutations are thought to be common events occurring in all sites of the tumor, and branch or subclonal mutations are thought to be responsible of the heterogeneity of tumors [63, 64]. This important classification has been linked to therapy outcomes in clinics [63] and the subject is extensively reviewed in [65,66,67].
Association of intra-tumor heterogeneity with poorer clinical outcomes has been documented in relapsed MGMT-deficient glioblastoma multiforme (GBM) following alkylator therapy [68] and relapsed acute myeloid leukemia (AML) following exposure to DNA-damaging agents [69]. Gillies et al. have suggested that cancer cytotoxics may contribute to tumor evolution by adversely changing the microenvironment, facilitating, hence, small phenotypic changes to result in large variations in fitness [70]. A study by Shah et al. on a case of advanced invasive lobular carcinoma of the breast illustrated the spatial and temporal dynamics of intra-tumor heterogeneity as 19 non-synonymous mutations have been detected in the metastases that were not present in the primary tumor diagnosed 9 years earlier [71]. This study is an example among many, illustrating the dynamic and often unpredictable behavior of tumor cells.
A more recent work by Gao et al. studying intra-tumor heterogeneity in hepatocellular carcinoma (HCC) focused on 26 genes previously characterized as drivers in HCC and mapped them to the phylogenetic trees. All 10 studied cases showed a long trunk with multiple branches where half of the mutations mapped to the trunk and the other half mapped to the branches [72].
Such modeling of mutational events into trunk and branches would theoretically help predict treatment course and success in individual patients. However, the knowledge of trunk and branch mutations may not translate in all cases into druggable targets as seen in the case of HCC where trunk mutations were the result of loss-of-function mutations [73]. Moreover, if we consider other levels of tumor heterogeneity such as intra-metastatic and inter-metastatic, the challenge of conquering cancer using DNA status information of tumors becomes seriously complicated. This is because there is no rule in cancer except the rule of becoming a cancer, i.e. switching from normalcy to malignancy. On the other hand, a passenger mutation at a branch could become a driver mutation when the tumor is subjected to pressure selection of drugs, making, therefore, the distinction between driver and passenger mutations irrelevant in that regard. In addition, co-targeting trunk and branch DNA alteration will more likely bring us back to the starting point of resistance and recurrence through formation of new mutations or the rising of new mechanisms of resistance due to selection pressure of drugs.
On the sunny side of the mutational tree, however, DNA mutations retain their role as prognostic and/or predictive markers with important consequences for patient care and treatment. Knowing the trunk and branch mutational identity could lead to important improvement in cancer patient outcome and intercept resistance as it rises. Computational methods can be used to predict mutations and knowing the possible trajectory of the tumor, metastases could be delayed if not prevented and the entire tumor be put under control. Effective, rational combination treatment in this way may help overcome resistance mechanisms in the short run but unlikely to do so in the long run.
The Impact of DNA Mutations on Cancer Treatment: the Breast Cancer Example
Breast cancer is the most common cancer in women worldwide [74]. In 2017, an estimated 255,180 new cases of invasive breast cancer are expected to be diagnosed in women in the USA, along with 63,410 new cases of non-invasive (in situ) breast cancer [75].
On a histological level, breast cancer is a heterogeneous disease with a range of diverse subtypes [76], whereas on a molecular level, genetic sub-typing describes the following breast cancers; luminal A, luminal B, HER2-enriched, and basal-like breast cancer (BLBC) with the triple-negative breast cancer (TNBC) being a sub-group of BLBC that itself has six sub-types, reviewed in [77]. This molecular sub-grouping is important in order to diagnose and define the appropriate treatment options currently available to breast cancer patients.
In recent decades, breast cancer treatment has evolved to a more target-directed approach and the objective here is to underline, through an example in HER2-type breast cancer, the major steps followed in the development of targeted drugs to see how the attribution of driver/passenger roles to the DNA mutations observed in cancer has influenced our approach towards treatment. The chronological order in the development of the drugs listed below is not as important as to what drugs have been designed and used to treat HER2-type breast cancer.
An Initial Drug: Antibody to Inhibit HER2
The human epidermal growth factor receptor 2 (HER2) is a 185-kDa trans-membrane protein encoded by the erbB2 oncogene located on chromosome 17q21–22 [78]. HER2 molecular marker is over-expressed in approximately 15–25% of primary human breast cancers and is shown to be associated with poor clinical outcomes and aggressive tumor progression [78, 79]. Patients with breast cancer tumors over-expressing HER2 are classically treated with anti-HER2 targeted therapy [80]. Trastuzumab was the first monoclonal antibody used to treat tumors over-expressing HER2. This humanized antibody, which binds to an extracellular segment of the HER2 receptor leading to inhibition of the proliferation of tumor cells, was shown to improve survival of patients with breast cancer over-expressing HER2 whether used as an adjuvant therapy in early stage disease [81, 82] or in combination with chemotherapy during the metastatic stage of the disease [83]. Studies have shown that the monoclonal antibody trastuzumab that targets HER2-positive breast cancer tumors when used in combination with chemotherapy results in a 33% reduction in the risk of death among HER2-positive patients [84]. Although HER2 targeted therapy had been shown to be beneficial, several patients developed recurrences or progression of the disease due to trastuzumab resistance [85].
The Need for a New Drug: a New Antibody to Inhibit HER2
A new humanized monoclonal antibody called pertuzumab has been developed that binds to a different domain of the extracellular portion of the HER2 receptor than trastuzumab and blocks HER2 dimerization [86]. A double-blind, randomized phase III trial compared the safety and efficacy of pertuzumab with trastuzumab and docetaxel, and showed a median progression-free survival of 18.5 months. An overall survival and objective response rate of 80.2% that is higher than the placebo group, led to the approval of pertuzumab for treating HER2-positive breast cancer [87]. Pertuzumab is, however, not used as a single agent but in combination with chemotherapy in HER2-positive breast cancer, hindering, hence, its further exploitation as a mono-therapy (reviewed in [88]).
The Need for Drug Combination
When confronted with continued unsatisfactory results, the classical approach is to try treatment combination. The aim is to explore the theoretical advantage of using two HER2 targeted agents for more complete blockade of the HER2 signaling pathway. Results obtained when combining pertuzumab and trastuzumab have been suggested to be more efficient in the treatment of HER2-positive breast cancer than the use of a single anti-HER2 agent in both metastatic breast cancer and preoperatively, as neoadjuvant therapy [89]. A randomized phase III trial evaluated the efficacy of dual targeted trastuzumab and lapatinib (a small-molecule tyrosine kinase) treatments in early-stage HER2-positive breast cancer but has found no significant statistical advantage in invasive disease-free survival over single agent trastuzumab treatment alone in an 8000-patient population size [90, 91].
Looking for New Targets in the PI3K/Akt/mTOR Pathway
Still registering poor results in HER2 breast cancer therapy, other targets in the same pathway have been used such as phosphatidylinositol 3-kinases (PI3K) in order to develop new inhibitors. In HER2 over-expressing breast cancer resistant to trastuzumab, the PI3K/Akt/mTOR has been further exploited and studies indicate that inhibitors of this pathway can act synergistically with trastuzumab in resistant cells [92].
Literature shows that increased signaling via up-stream growth factor receptors, such as those in the GFR and IGFR1 families, PTEN mutations, and changes in the HER2 receptor, all play an important role leading to resistance to anti-HER2 agents. The major mechanism recorded for trastuzumab resistance appears, however, to be increased activation/signaling of PI3K/Akt [93]. Therefore, the combination of trastuzumab with an mTOR inhibitor has been shown to act synergistically to inhibit tumor proliferation, and the addition of trastuzumab to an mTOR inhibitor reduces the activity of the PI3K, MAPK and HER3 signaling pathways [94]. Several PI3K or Akt inhibitors are currently in clinical or preclinical studies but no agent is currently FDA-approved (reviewed in [95, 96]).
A Small-Molecule Tyrosine Kinase to Inhibit HER2
Small-molecule tyrosine kinase inhibitors such as lapatinib have also been developed to target HER2. This inhibitor binds to the ATP-binding pocket of the EGFR/HER2 protein kinase domain, preventing self-phosphorylation and subsequent activation [97]. Several small-molecule inhibitors of the kinases are presently undergoing phase trials (reviewed in [98]) and the efficiency of such therapeutic agents remains unclear.
Summarizing the approach followed for HER2 breast cancer treatment shows this sequence of events: (1) development of a first inhibitor to target a major molecule in a given pathway characterizing that tumor (anti-HER2 antibody); (2) registering resistance to the first type of inhibitors leads to the design of a new inhibitor of the same nature (new anti-HER2 antibody); (3) unsatisfactory results lead to drug combination; (4) partial or unsatisfactory results lead to search for new targets down-stream the same metabolic pathway (PI3K/Akt/mTOR inhibitors). The vicious cycle goes on and on, mainly when we consider other types of cancer affecting the breast and also other organs.
The questions asked here are: why the first/second antibodies or the small-molecule kinase inhibitor did not work as expected and give a durable positive response? And why treatment combinations did not work either? To answer the why question seems here more important than answering the how question. Though gaining knowledge on how resistance arises is important, the usage of this information is unlikely to lead to breast cancer eradication. Inhibiting the usage of a major tool (here, HER2 over-expression) on which cancer cells rely for their growth, has not stopped cancerous cells from growing or coming back. In other words, where is the specificity of the targeting inhibitors used to target the HER2-type of breast cancer?
The answer perhaps lies in the fact that the targets used to generate such inhibitors are not the drivers as thought. Were these DNA mutations the drivers, then the inhibition of their gene product should have resulted in positive and durable response. This was not the case, not only in HER2 breast cancer elucidated here but also in other types of breast cancer such as TNBC where PARP targeting has also been met with resistance [99].
Or, could these DNA mutations be just symptoms of transformation instead of its driving cause? And while what really drives cancer is not targeted with these targeted therapies, then cancerous cells need only time to figure out, when confronted with a non-specific drug, how to go over the fence or cross the road and continue to grow, using different molecular tools to either show resistance or relapse a few months later.
Conclusions
Tumor DNA sequencing data has shown that DNA mutations in cancer cells are multiple and heterogeneous. Today, enough sequencing data have been generated showing that DNA mutations thought to be drivers in HER2-type of breast cancer or EGFR-positive NSCLC have not resulted in long-lasting positive response when targeted with different drugs. These numerous DNA mutations detected in cancerous cells in general could be regarded as symptoms or consequences of transformation suggesting that the driver in cancer may not be a particular DNA mutation or a particular spectrum of DNA mutations. The non-randomness seen in the pattern of DNA mutations in cancer cells may not translate into a causative role. This non-randomness could reflect the patterns of normal cells which are often hijacked in transformed cells since cancer cells are unable to create de novo metabolic pathways [33]. Rules and regulations governing cell division exist in normal but not in cancerous cells. As a result, cancerous cells have shown a multitude of pathways they can subdue to grow into many histological and molecular types and sub-types as seen here in breast cancer. Heterogeneity in tumors reflects this unruly behavior in cancer cells which, at the same time, reflects their remarkable adaptation and changing character.
Moreover, mutations do occur in DNA but without causing cancer. More importantly though and in order to prevent transformation of multi-cellular organisms and preserve their nature, evolution constraints may be in action and serve as a barrier to oppose all transforming changes capable of affecting a species as a whole. Spontaneous abortions seen in humans could be one of such constraints. On the other hand, a cancerous cell is a normal cell that has been transformed and tumor masses look nothing like their normal tissues from which they took root and such outcome is prevented from happening at a level of species. Through that evolutionary control, the material bearing genetic information—characteristic of each species—is protected in order to prevent species from being transformed as a consequence.
In addition to the drugs listed above, resistance registered with PARP inhibitors is another well-documented and clear-cut example showing how our interpretation of the knowledge gained in the field of DNA mutations has influenced our therapeutic approach. Cancerous cells have a malignant approach and when they mutate BRCA1/2, they disempower homologous recombination which is an error-free repair pathway, to open the door to an error-prone repair pathway such as NHEJ to take place. Blocking the PARP pathway used by cancerous cells could translate in an increase in the aggressiveness of the tumors treated with PARPi as they become more skilled, should these tumors resort to an error-prone repair pathway. Moreover, it has been reported that the elevated frequency of micro-homology-mediated insertions-deletions observed in BRCA1 or BRCA2 mutant cancers are thought to reflect the usage of alternative methods of DSB repair in these cancers [100] and has been reviewed in [101, 102].
As stated by Gonzalez-Angulo et al., breast cancer resistance to therapy is not only common but expected as systemic agents are active at the beginning of therapy in 90% of primary breast cancers and 50% of metastases. However, after a variable period of time, progression occurs [100]. The time needed for cancerous cells before they show full resistance or before relapsing may depend on the tumor’s microenvironment and also on how far the tumor has grown and how many tools it has at hand, i.e. how many genes it had already mutated. Focusing on how resistance arises engaged us in a vicious circle, with resistance leading to new drugs for new targets, leading to new combinations of drugs, but without achieving better and long-lasting clinical outcomes.
The cancer trilogy we came to is obvious in its following sequence: initial drug; tumor-resistance/relapse; drug/treatment-combination. This is also seen in clinics in all types of cancers with very few exceptions. Moreover, it is important to mention that we cannot make right something that is fundamentally wrong no matter how hard we try, and when a problem persists, it often calls for a paradigm shift. Besides the numerous breakthroughs in genome sequencing results, our understanding of cancer as a disease remains poor. The focus on the tumor mass with the goal of downsizing it should shift to the single-cell level with the goal of stopping each and every cancerous cell from making its first round of cell division regardless of its genetic repertoire. How a cancerous cell divides should shift to what makes a cancerous cell divide without control. How a cancerous cell divides and grows has led us so far to develop inhibitors to the tools (symptoms) these cancerous cells rely on such as an over-expressed molecule. What makes a cancerous cell create those tools, use them intelligently to defy our inhibiting strategies is what needs more attention.
DNA mutations, considered drivers in breast cancer and other types of cancer, offer, however, valuable information as biomarkers which can influence diagnosis and risk assessment. The aim of using DNA profiling is to identify tumors which most likely will respond to currently used agents or combination of agents. However, in clinical practice, oncologists are often challenged in how to define the therapeutic sequence that is most efficient and to use DNA mutation information to reach an optimum positive response. Seeking optimization of existing treatment remains, therefore, important. In this regard, it is essential to underlie a study conducted on high-grade serous ovarian cancer (HGSOC) supporting the benefit of PARP targeted therapy [103]. The study conducted on three patients showed somatic disruption of BRCA1/2 in all three patients at diagnosis, followed by subsequent BRCA1 recovery upon progression by copy-number gain and/or up-regulation of the remaining functional allele in two patients. The third patient with ongoing response (>7 years) had a tumor at diagnosis with bi-allelic somatic deletion and loss-of-function mutation, thereby lacking a functional allele for recovery of BRCA1 activity. This highlights a potential cure for such a patient thanks to the usage of sequencing data information.
On the other hand, a more recent meta-analysis study on pancreatic cancer failed to show a significant improvement of targeted therapy in comparison with chemotherapy, outlining the challenges still imposed by targeted therapy [104]. Resistance to currently used therapies will likely continue to unfold as long as the real driver in cancer is not targeted. Cancer growth is an evolutionary process and this evolution plays a key role in both the development of tumors and their response to therapy. Hirata and Sahai, discussed how therapeutic strategies directly alter the composition or function of the tumor microenvironment and in doing so, they further alter the selective pressures to which cancer cells are exposed (reviewed in [105]).
Finally, the objective in cancer therapy should not be limited to improving the overall survival of cancer patients but rather to cure all cancer patients regardless of the genetic characteristics of their tumors. The complexity of cancer as a disease and the many challenges encountered today with resistance and relapse call for a new definition of cancer. Cancer may not be primarily a genetic disease, insinuating DNA changes as causal events as described in literature. Cancer could rather be described as a disease caused but something still unknown but characterized, at a cellular level, by DNA alterations needed to reprogram a normal cell in order to develop a malignant behavior.
Future cancer research should focus on identifying the initial event responsible for the switch from normalcy to malignancy and what switches a cell from normalcy to malignancy is the cause of cancer. Targeting the cause of cancer will inevitably result in a cancer cure. Working towards that end and before putting a finger on the cause of cancer, it is imperative to first explain important issues in malignancy, reconciling these genetic manifestations into a comprehensive model. Cancer hallmarks described by Hanahan and Weinberg [106] provide an elegant framework for the development of cancer and offer a solid basis in this endeavor. Moreover, the classical multi-stage model of carcinogenesis may explain tumor evolution but does not explain tumor initiation thought here unlikely to lie on DNA sequences. Caiado et al. have recently reported that four major interdependent non-genetic factors are suggested to critically contribute to the overall variability of tumor cells in all types of cancer: epigenetic regulation, gene expression stochasticity, cellular differentiation hierarchies and tumor microenvironment [107]. Therefore, the time is ripe to go a step farther and move cancer research in a fundamentally new direction. The next few years could see a dramatic change in cancer care and treatment. While trying novel venues, targeted therapy remains on the scene and can bring newer and important improvements in the lives of breast cancer patients and also in other types of cancer; however, it is unlikely to result in breast cancer eradication depicted here.
References
Butler T, Maravent S, Boisselle J, et al. A review of 2014 cancer drug approvals, with a look at 2015 and beyond. P&T. 2015;40(3):191–205.
Housman G, Byler S, Heerboth S, et al. Drug resistance in cancer: an overview. Cancers. 2014;6:1769–92. doi:10.3390/cancers6031769.
Tian F, Zhao J, Fan X, et al. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. J Thorac Dis. 2017;9(1):42–53. doi:10.21037/jtd.2017.01.04.
Nulton TJ, Olex AL, Dozmorov M, et al. Analysis of the cancer genome atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget. 2017;8(11):17684–99. doi:10.18632/oncotarget.15179.
Cancer Genome Anatomy Project (CGAP) Cancer Genome Characterization Initiative (CGCI). Cgap.nci.nih.gov. As retrieved on Sept 14, 2013. doi:10.1146/annurev-genom-082509-141532.
Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics of cancer: genome sequencing and beyond. Annu Rev Genom Hum Genet. 2011;12:407–30.
Futreal PA, Lachlan C, Mhairi M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–83. doi:10.1038/nrc1299.
Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):8–9.
Muller FL, Colla S, Aquilanti E, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488(7411):337–42.
Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–6.
Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–8.
Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Straton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:7239. doi:10.1038/nature07943.
Jones SJM, Laskin J, Li YY, et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010;11(8):R82. doi:10.1186/gb-2010-11-8-r82.
Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111ra121. doi:10.1126/scitranslmed.3003161.
Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94.
Arteaga C. Targeting HER1/EGFR: a molecular approach to cancer therapy. Semin Oncol. 2003;30:3–14.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib therapy. N Engl J Med. 2004;350:2129–39.
Zhang H. Three generations of epidermal growth factor receptor tyrosine kinase inhibitors developed to revolutionize the therapy of lung cancer. Drug Des Dev Ther. 2016;10:3867–72.
Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92.
Oxnard GR, Thress K, Paweletz C, et al. Mechanisms of acquired resistance to AZD9291 in EGFR T790M positive lung cancer. J Thorac Oncol. 2015;10:1736–44.
Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74.
Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–6.
Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63.
Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6.
Ding L, Kim M, Kanchi KL, et al. Clonal Architectures and driver mutations in metastatic melanomas. PLoS One. 2014;9(11):e111153. doi:10.1371/journal.pone.0111153.
Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29:5545–55.
Mar VJ, Liu W, Devitt B, et al. The role of BRAF mutations in primary melanoma growth rate and survival. Br J Dermatol. 2015;173(1):76–82.
Kumar R, Angelini S, Snellman E, et al. BRAF mutations are common somatic events in melanocytic nevi. J Invest Dermatol. 2004;122(2):342–8.
Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J Natl Cancer Inst. 2013;105(12):917–9.
Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi:10.1038/ng.2983.
Adjiri A. Identifying and targeting the cause of cancer is needed to cure cancer. Oncol Ther. 2016;4:17. doi:10.1007/s40487-015-0015-6.
Beerenwinkel N, Antal T, Dingli D, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3(11):e225. doi:10.1371/journal.pcbi.0030225.
Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115–30. doi:10.1016/j.ccr.2007.07.004.
Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the non-canonical NF-kB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–44.
Hafner C, López-Knowles E, Luis NM, et al. Oncogenic PIK3CA mutations occur in epidermal nevi and seborrheic keratoses with a characteristic mutation pattern. Proc Natl Acad Sci USA. 2007;104(33):13450–4.
Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19–20.
Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127(1):179–82.
Poynter JN, Elder JT, Fullen DR, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16(4):267–73. doi:10.1097/01.cmr.0000222600.73179.f3.
Raaijmakers MIG, Widmer DS, Narechania A, et al. Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget. 2016;7(47):77163–74.
Daya-Grosjean L, Dumaz N, Sarasin A. The specificity of p53 mutation spectra in sunlight induced human cancers. J Photochem Photobiol B. 1995;28(2):115–24.
Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat Res. 2005;571(1–2):43–56.
Nunney L, Muir B. Peto’s paradox and the hallmarks of cancer: constructing an evolutionary framework for understanding the incidence of cancer. Philos Trans R Soc Biol. 2015;370:20150161. doi:10.1098/rstb.2015.0161.
Futreal PA, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–2.
Lancaster JM, Wooster R, Mangion J, et al. BRCA2 mutations in primary breast and ovarian cancers. Nat Genet. 1996;13:238–40.
Olivier M, Goldgar DE, Sodha N, et al. Li–Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 2003;63(20):6643–50.
Rème T, Travaglio A, Gueydon E, et al. Mutations of the p53 tumour suppressor gene in erosive rheumatoid synovial tissue. Clin Exp Immunol. 1998;111(2):353–8.
Firestein GS, Echeverri F, Yeo M, et al. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci USA. 1997;94(20):10895–900.
Malkin D. Li–Fraumeni syndrome. Genes Cancer. 2011;2(4):475–84. doi:10.1177/1947601911413466.
Fearon ER. Human cancer syndromes: clues to the origin and nature of cancer. Science. 1997;278(5340):1043–50.
Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786(1):32–40.
Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–67.
Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modeling of the human disease. Nat Rev Cancer. 2010;10(2):102–15.
Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13.
Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7.
Fujiwara T, Bandi M, Nitta M, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–7.
Weaver BA, Silk AD, Montagna C, et al. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36.
Silk AD, Zasadil LM, Holland AJ, et al. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci USA. 2013;110(44):E4134–41.
Menasha J, Levy B, Hirschhorn K. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med. 2005;7(4):251–63.
Jacqueline C, Biro PA, Beckmann C, et al. Cancer: a disease at the crossroads of trade-offs. Evol Appl. 2016;10(3):215–25. doi:10.1111/eva.12444 (eCollection 2017).
Venkatesan S, Birkbak NJ, Swanton C. Constraints in cancer evolution. Biochem Soc Trans. 2017;45(1):1–13. doi:10.1042/BST20160229.
Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Yap TA, Gerlinger M, Futreal PA, et al. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi:10.1126/scitranslmed.3003854.
Swanton C. Intratumour heterogeneity: evolution through space and time. Cancer Res. 2012;72(19):4875–82. doi:10.1158/0008-5472.CAN-12-2217.
Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;21(6):1258–66. doi:10.1158/1078-0432.CCR-14-1429.
Turajlic S, McGranahan N, Swanton C. Inferring mutational timing and reconstructing tumour evolutionary histories. Biochem Biophys Acta. 2015;1855:264–75.
McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8.
Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–10.
Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12(7):487–93.
Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–13.
Gao Q, Wang ZC, Duan M, et al. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology. 2017;152:232–42.
Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21:1951–61.
McGuire A, Brown JA, Malone C, et al. Effects of age on the detection and management of breast cancer. Cancers. 2015;7(2):908–29. doi:10.3390/cancers7020815.
http://www.breastcancer.org/symptoms/understand_bc/statistics. Last modified on Jan 10, 2017 at 10:51 AM.
Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi:10.1016/j.molonc.2010.04.004.
Narayanan R, Dalton JT. Androgen receptor: a complex therapeutic target for breast cancer. Cancers. 2016;8:108. doi:10.3390/cancers8120108.
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12.
Bacus SS, Zelnick CR, Plowman G, Yarden Y. Expression of the erbb-2 family of growth factor receptors and their ligands in breast cancers. Implication for tumor biology and clinical behavior. Am J Clin Pathol. 1994;102:S13–24.
Browne BC, O’Brien N, Duffy MJ, Crown J, O’Donovan N. HER-2 signaling and inhibition in breast cancer. Curr Cancer Drug Targets. 2009;9(3):419–38.
Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–83.
Yin W, Jiang Y, Shen Z, et al. Trastuzumab in the adjuvant treatment of HER2-positive early breast cancer patients: a meta-analysis of published randomized controlled trials. PLoS One. 2011;6(6):e21030. doi:10.1371/journal.pone.0021030.
Perez EA, Romond EH, Suman VJ, et al. Original report: 4-year follow- up of trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSAB B-31. J Clin Oncol. 2011;29:3366–73.
Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84.
Vu T, Claret FX. Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol. 2012;2:62. doi:10.3389/fonc.2012.00062.
Baselga J, Gelmon KA, Verma S, et al. Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol. 2010;28:1138–44.
Boix-Perales H, Borregaard J, Jensen KB, et al. The european medicines agency review of pertuzumab for the treatment of adult patients with HER2-positive metastatic or locally recurrent unresectable breast cancer: summary of the scientific assessment of the committee for medicinal products for human use. Oncologist. 2014;19:766–73.
Hubalek M, Brantner C, Marth C. Role of pertuzumab in the treatment of HER2-positive breast cancer. Breast Cancer Targets Ther. 2012;4:65–73.
Ahn ER, Vogel CL. Dual HER2-targeted approaches in HER2-positive breast cancer. Breast Cancer Res Treat. 2012;131(2):371–83. doi:10.1007/s10549-011-1781-y (Epub 2011 Sep 29).
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–72.
Piccart-Gebhart MJ, Holmes AP, Baselga J, et al. First results from the phase III ALTTO trial (BIG 2-06; NCCTG [alliance] N063D) comparing 1 year of anti-HER2 therapy with lapatinib alone (L), trastuzumab alone (T), their sequence (T → L), or their combination (T + L) in the adjuvant treatment of HER2-positive early breast cancer (EBC). J Clin Oncol. 2014;32 Suppl 5s:Abstract LBA4.
Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–9.
Nahta R, O’Regan RM. Evolving strategies for overcoming resistance to HER2-directed therapy: targeting the PI3K/Akt/mTOR pathway. Clin Breast Cancer. 2010;10(Suppl 3):S72–8.
Miller TW, Forbes JT, Shah C, et al. Inhibition of mammalian target of rapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2-overexpressing cancer cells. Clin Cancer Res. 2009;15(23):7266–76.
Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66. doi:10.1177/1758834014530023.
Lee JJX, Loh K, Yap YS. PI3K/Akt/mTOR inhibitors in breast cancer. Cancer Biol Med. 2015;12:342–54. doi:10.7497/j.issn.2095-3941.2015.0089.
Konecny GE, Pegram MD, Venkatesan N, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–9.
Schroeder RL, Stevens CL, Sridhar J. Small molecule tyrosine kinase inhibitors of ErbB2/HER2/Neu in the treatment of aggressive breast cancer. Molecules. 2014;19:15196–212. doi:10.3390/molecules190915196).
Benafif S, Hall M. An update on PARP inhibitors for the treatment of cancer. Oncotargets Ther. 2015;8:519–28.
Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22.
Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93.
Kelley MR, Logsdon D, Fishel ML. Targeting DNA repair pathways for cancer treatment: what’s new? Future Oncol. 2014;10(7):1215–37. doi:10.2217/fon.14.60.
Lheureux S, Bruce JP, Burnier JV, et al. Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly (ADP-ribose) polymerase inhibition. J Clin Oncol. 2017;. doi:10.1200/JCO.2016.71.3677.
Ottaiano A, Capozzi M, De Divitiis C, et al. Gemcitabine mono-therapy versus gemcitabine plus targeted therapy in advanced pancreatic cancer: a meta-analysis of randomized phase III trials. Acta Oncol. 2017;56(3):377–83. doi:10.1080/0284186X.2017.1288922 (Epub 2017 Feb 17).
Hirata E, Sahai E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med. 2017;. doi:10.1101/cshperspect.a026781.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Caiado F, Silva-Santos B, Norell H. Intra-tumour heterogeneity—going beyond genetics. FEBS J. 2016;283(12):2245–58. doi:10.1111/febs.13705.
Acknowledgements
No funding or sponsorship was received for this study or publication of this article. The named author meets the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, takes responsibility for the integrity of the work as a whole, and has given final approval for the version to be published.
Disclosures
A. Adjiri has nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not involve any new studies of human or animal subjects performed by any of the authors.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced content
To view enhanced content for this article go to http://www.medengine.com/Redeem/B028F06073B447FA.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Adjiri, A. DNA Mutations May Not Be the Cause of Cancer. Oncol Ther 5, 85–101 (2017). https://doi.org/10.1007/s40487-017-0047-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40487-017-0047-1