Abstract
Background
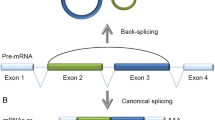
Circular RNAs (circRNAs) from back-spliced exon(s) are characterized by the covalently closed loop feature with neither 5′ to 3′ polarity nor polyadenylated tail. By using specific computational approaches that identify reads mapped to back-splice junctions with a reversed genomic orientation, ten thousands of circRNAs have been recently re-identified in various cell lines/tissues and across different species. Increasing lines of evidence suggest that back-splicing is catalyzed by the canonical spliceosomal machinery and modulated by cis-elements and trans-factors.
Results
In this mini-review, we discuss our current understanding of circRNA biogenesis regulation, mainly focusing on the complex regulation of complementary sequences, especially Alus in human, on circRNA formation.
Conclusions
Back-splicing can be significantly facilitated by RNA pair formed by orientation-opposite complementary sequences that juxtapose flanking introns of circularized exon(s). RNA pair formed within individual introns competes with RNA pair formed across flanking introns in the same gene locus, leading to distinct choices for either canonical splicing or back-splicing. Multiple RNA pairs that bracket different circle-forming exons compete for alternative back-splicing selection, resulting in multiple circRNAs generated in a single gene locus.

Article PDF
Similar content being viewed by others
References
Chen, L. L. (2016) The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol., 17, 205–211
Chen, L. L. and Yang, L. (2015) Regulation of circRNA biogenesis. RNA Biol., 12, 381–388
Lasda, E. and Parker, R. (2014) Circular RNAs: diversity of form and function. RNA, 20, 1829–1842
Yang, L. (2015) Splicing noncoding RNAs from the inside out. WIREs RNA, 6, 651–660
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., Zhu, S., Yang, L. and Chen, L. L. (2013) Circular intronic long noncoding RNAs. Mol. Cell, 51, 792–806
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., Marzluff, W. F. and Sharpless, N. E. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19, 141–157
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., Maier, L., Mackowiak, S. D., Gregersen, L. H., Munschauer, M., et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495, 333–338
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. and Brown, P. O. (2013) Cell-type specific features of circular RNA expression. PLoS Genet., 9, e1003777
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. and Brown, P. O. (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One, 7, e30733
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L. and Yang, L. (2014) Complementary sequence-mediated exon circularization. Cell, 159, 134–147
Yang, L., Duff, M. O., Graveley, B. R., Carmichael, G. G. and Chen, L. L. (2011) Genomewide characterization of nonpolyadenylated RNAs. Genome Biol., 12, R16
Yin, Q. F., Chen, L. L. and Yang, L. (2015) Fractionation of nonpolyadenylated and ribosomal-free RNAs from mammalian cells. Methods Mol. Biol., 1206, 69–80
Zhang, Y., Yang, L. and Chen, L. L. (2016) Characterization of Circular RNAs. Methods Mol. Biol., 1402, 215–227
Chen, L. L. and Yang, L. (2017) ALUternative regulation for gene expression. Trends Cell Biol., 27, 480–490
Hansen, T. B., Veno, M. T., Damgaard, C. K. and Kjems, J. (2016) Comparison of circular RNA prediction tools. Nucleic Acids Res., 44, e58
Zhang, X. O., Dong, R., Zhang, Y., Zhang, J. L., Luo, Z., Zhang, J., Chen, L. L. and Yang, L. (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res., 26, 1277–1287
Jeck, W. R. and Sharpless, N. E. (2014) Detecting and characterizing circular RNAs. Nat. Biotechnol., 32, 453–461
Dong, R., Ma, X. K., Chen, L. L. and Yang, L. (2016) Increased complexity of circRNA expression during species evolution. RNA Biol., 1–11. doi: 10.1080/15476286.2016.1269999
Ivanov, A., Memczak, S., Wyler, E., Torti, F., Porath, H. T., Orejuela, M. R., Piechotta, M., Levanon, E. Y., Landthaler, M., Dieterich, C., et al. (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep., 10, 170–177
Rybak-Wolf, A., Stottmeister, C., Glazar, P., Jens, M., Pino, N., Giusti, S., Hanan, M., Behm, M., Bartok, O., Ashwal-Fluss, R., et al. (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell, 58, 870–885
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., Celniker, S. E., Graveley, B. R. and Lai, E. C. (2014) Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep., 9, 1966–1980
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., Evantal, N., Memczak, S., Rajewsky, N. and Kadener, S. (2014) circRNA biogenesis competes with premRNA splicing. Mol. Cell, 56, 55–66
Starke, S., Jost, I., Rossbach, O., Schneider, T., Schreiner, S., Hung, L. H. and Bindereif, A. (2015) Exon circularization requires canonical splice signals. Cell Rep., 10, 103–111
Zhang, Y., Xue,W., Li, X., Zhang, J., Chen, S., Zhang, J. L., Yang, L. and Chen, L. L. (2016) The biogenesis of nascent circular RNAs. Cell Rep., 15, 611–624
Liang, D. and Wilusz, J. E. (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev., 28, 2233–2247
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody,M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh,W., et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921
Chen, L. L., DeCerbo, J. N. and Carmichael, G. G. (2008) Alu element — mediated gene silencing. EMBO J., 27, 1694–1705
Guarnerio, J., Bezzi, M., Jeong, J. C., Paffenholz, S. V., Berry, K., Naldini, M. M., Lo-Coco, F., Tay, Y., Beck, A. H. and Pandolfi, P. P. (2016) Oncogenic role of fusion-circRNAs derived from cancerassociated chromosomal translocations. Cell, 165, 289–302
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., Roslan, S., Schreiber, A.W., Gregory, P. A. and Goodall, G. J. (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell, 160, 1125–1134
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S. and Wilusz, J. E. (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev., 29, 2168–2182
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., Davey, N. E., Humphreys, D. T., Preiss, T., Steinmetz, L. M., et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406
He, C., Sidoli, S., Warneford-Thomson, R., Tatomer, D. C., Wilusz, J. E., Garcia, B. A. and Bonasio, R. (2016) High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell, 64, 416–430
Li, X., Liu, C. X., Xue,W., Zhang, Z., Jiang, S., Yin, Q. F., Wei, J., Yao, R.W., Yang, L. and Chen, L. L. (2017) Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell, http://doi.org/10.1016/j.molcel.2017.05.023
Aktaş, T., Avşar Ilık, İ., Maticzka, D., Bhardwaj, V., Pessoa Rodrigues, C., Mittler, G., Manke, T., Backofen, R. and Akhtar, A. (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature, 544, 115–119
Acknowledgments
We are grateful to L.-L.C. for critical reading of this manuscript. We apologize to authors whose work could not be cited here owing to space/ content limitations. Our work is supported by grants 2014CB910601 from MOST and 91540115 and 31471241 from NSFC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, Q., Wang, Y. & Yang, L. Multifaceted roles of complementary sequences on circRNA formation. Quant Biol 5, 205–209 (2017). https://doi.org/10.1007/s40484-017-0112-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40484-017-0112-7