Abstract
Purpose of Review
The global COVID-19 pandemic has claimed millions of lives and harmed hundreds of millions more. Amidst this crisis, scientists have used multi-omics to understand and combat the virus. The purpose of this review is to provide the latest and most impactful work in COVID-omics.
Recent Findings
Multi-omics has identified risk-stratification criteria to predict viral severity among COVID-19 patients. Omic methods have also unlocked targetable biomarkers in viral pathways and enabled public health agencies to curb transmission by genomic tracing. Transplant researchers have used multi-omics to assess the safety of transplanting organs from COVID-positive donors, and whether patient immunosuppression regimens should be maintained. Lastly, maximizing multi-omic impact by nurturing future collaborations between mutli-omic labs and public health agencies and pharmaceutical companies will be critical in successfully facing the next pandemic.
Summary
This review focuses on contributions within the field of COVID-omics, including patient risk stratification and viral pathway analysis, genomic public health surveillance, and transplant clinician recommendations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In transplantation as well as pandemic relief, time is a priceless resource. One month after the first reported cases in Wuhan, China, the WHO declared COVID-19 an international public health emergency. In less than a year, over 30 million cases were reported globally [1]. The unprecedented strain of this highly contagious virus upon healthcare delivery systems saw crucial shortages of beds and ventilators for patients, as well as PPE for providers [2, 3]. As of 2023, millions of lives have been lost due to the virus [4]. The world has yet to recover, with nations reeling from impacted economies, separated families, and traumatized communities whose health continues to deteriorate from long COVID and prolonged isolation [5, 6].
When it comes to eliminating preventable deaths, time remains one of our greatest defenses. Knowing how a virus operates, behaves, or differentially impacts individuals may allow us to stop transmission before it occurs, and to better care for those affected. Toward this end, COVID-19 has become the subject of intense multi-omic research [7]. Multi-omics is a systems biology approach that applies mathematical modeling and machine learning to large-scale molecular data generation. Mapping diverse molecular interactions allows scientists to examine intricate layers of biological connections to yield translational insights [8]. Early indicators of disease, a central concern of precision medicine and biomedical research generally, have been successfully captured by multi-omic research across medical specialties [9].
This review surveys the latest and most impactful work in COVID-19 and transplantation multi-omics. The first section assesses the generation of patient risk stratification criteria and therapeutic targets within pathways from multi-omics methods. The second explores recent research in genomics and patient transmission prediction, as well as groundbreaking work regarding genetic susceptibility to the virus and the ethics of genomic-based clinical decision-making. The third and final section examines the intersection of multi-omics and organ transplantation in the era of COVID-19. The overall aim of this review is to update the transplant community on the rise and trajectory of pandemic research in the multi-omics field, and summarize the promise that relevant methodologies hold for bedside applications.
Diagnosis and Targeting: COVID-Omics for Clinical Interventions
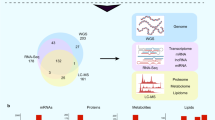
Multi-omics is highly useful in revealing predictive markers for disease severity and progression. Studies have grown to involve study cohorts in the tens of thousands of patients, often with robust external validation across multiple healthcare sites (Fig. 1). Delafiori enrolled a cohort of 815 patients from three Brazilian pandemic epicenters and used machine learning to develop a model for assessing viral risk severity. Using metabolomic data from patient plasma samples, the researchers identified seven biomarkers indicating severe COVID-19 infection (among them, deoxyguanosine/adenosine, N-stearoyl valine, and sterol lipid derivatives), and nineteen indicative of mild risk (including four glycerophospholipids, two glycerolipids, and one peptide) [10]. Additionally, Zhou validated genome-wide whole-blood transcriptomic data in a multicenter cohort of over 1200 patients [11]. By incorporating machine learning techniques with omic data, the researchers developed a prediction algorithm capable of stratifying patients into in-hospital mortality risk categories (divided by 15-day, 30-day, and 45-day stays) based on the interplay of platelet, neutrophil, lymphocyte, and hemoglobin fluctuations.
In addition to patient risk stratification, associative studies within multi-omics have powered insights into COVID-19 viral pathways. Early proteomic analyses gave insight into the pathogenesis of COVID-19. Critical increases of biomarkers such as fibrinogen alpha (FGA) and beta (FGB) within the fibrinolytic cascade were present in the sera of patients with severe COVID-19 [12•]. This gave the building blocks to the hypercoagulability seen in COVID-19 patients and drove landmark critical trials in COVID-19 evaluating the use of anticoagulation in the setting of this illness. In addition, organ autopsy samples in COVID-19 patients showed dysregulation in multiple organs and systemic hyperinflammation [13]. Further omic studies pointed out biomarker fluctuations and unusual immune cell activity, such as hyperactivation signatures in neutrophils and NK cells, which explained the exaggerated host immune response and underscored potential targets and settings for therapeutic intervention [14]. Stephenson (2021) offers an especially thorough analysis of the coordinated immune response through a mass analysis of peripheral blood mononuclear cells from patients with varying severities of COVID-19 [15•]. They identified a deteriorating COVID-19 signature as involving an increase of lymphocytes and monocytes, and found that CCL4, CXCL10, interleukin (IL)-7 and IL-1α in particular were associated with severe and critical disease.
Among such associative work, Overmyer stands out as a highly cited transcriptomic, proteomic, metabolomic, and lipidomic analysis of blood samples from COVID-19 patients [13]. The authors identified and correlated over 17,000 biomolecules with patient outcomes and clinical data to generate an interactive web-based database (covid-omics.app), which is now freely available for exploration. Of these biomarkers, the authors note that citrate, an anticoagulant; plasmenyl-PCs, which regulate inflammation; and pGSN, another inflammation inhibitor, are each shown to decrease with COVID-19 severity. However, these molecules can be clinically supplemented, potentially offering an immediate therapeutic solution for at-risk patients. Additionally, they illustrate the utility of their omic data by developing a robust machine-learning-trained model for COVID-19 severity, with severity defined by criteria such as hospitalization and number of days spent on ventilatory support. These metrics were combined with other existing severity indices, such as the Charlson comorbidity index score and sequential organ failure assessment (SOFA) score, to comprehensively evaluate disease progression.
More recent studies are similarly invested in converting associative data to clinical interventions. One longitudinal study first used transcriptomics to uncover 5915 differentially expressed genes in COVID-19 patients, building a disease framework encompassing four distinct clinical stages based on total time of hospitalization (asymptomatic; less than 10 days; over 20 days; and deceased) [14]. Their subsequent metabolic analysis revealed hyperactivation of plasmablasts and a shift toward amino acid metabolism over glycolysis, suggesting a link between such immunometabolic features and fatal outcomes. In another metabolomic study, over 70 metabolites were differentially expressed and found to be altered by severe COVID-19 [15, 16]. This study identified anthranilic acid as participating in the kynurenine pathway that may be inhibited to prevent patient immunosuppression. Further metabolomic studies have investigated therapeutic interventions in glucose metabolism, the urea cycle, and amino acid metabolism in COVID-19 patients [17].
As illustrated above, multi-omics offer numerous advantages for clinicians. Not only do we exist in a technological age of big data: the analytical methods are becoming cheaper every year [18]. Not only are viral outcomes and pathways elucidated at the molecular level: COVID-omic researchers are using omic data to advance the conversation and expand their insights beyond their immediate network. Free online databases, such as the multi-omic dataset COVINET, have bloomed to highlight hotspots that might be targeted by existing drugs, sharing a wealth of insight with the scientific community [16]. When such studies take into consideration pharmaceutical applications, they open up the possibilities for researchers to identify FDA-approved drugs suitable for repurposing as COVID-19 medications [19].
However, there are notable drawbacks to clinical use of multi-omic data for treating COVID-19. First, the use of machine learning algorithms in omic research for patient risk stratification might result in overgeneralized clinical recommendations [20]. Clinicians must integrate these broad indicators with patient-specific recommendations in order to maximize the benefits of omic knowledge. Furthermore, the matter of execution remains. Despite the breadth of omic studies in circulation during the height of the crisis, the disconnect between academic research labs and pharmaceutical pipelines has distanced multi-omics knowledge from impacting real-time drug development. Until such a gap can be bridged, multi-omics’ greatest impact might reside within the realm of clinical decision-making.
Tracking the Virus: Genomics and Transmission
COVID-19 is exceedingly contagious. Transmitted largely by contact and droplet forms, its superspreading infectivity resulted in a tenfold multiplication over 10 days, from a few initial positive cases [21]. Protecting human life by containing its spread cannot rely on diagnostic markers alone: as Larsen notes, “hospitalizations lag infections by weeks and do not report on people with mild or asymptomatic disease” [22]. The situation is further complicated by social distancing restrictions and resource shortages. Barriers to accessing testing stations or kits further impedes diagnostics and skews the total number of cases. As the first supervirus in the era of omics—and genomics in particular—COVID-19 knowledge has benefited from approaches not possible during past outbreaks such as the SARS epidemic [23].
Genomics, which focuses on the structure and evolution of genomes, is able to take advantage of mutations that naturally occur during the viral replication cycle, and create diversified lineages. Although COVID-19 and its fellow coronaviruses possess certain mechanisms to preserve genomic fidelity, structural and molecular variations nonetheless provided genomic researchers critical insight into the origins and transmission dynamics of the virus [24]. Viral genomes obtained from nine COVID-19 patients, eight of whom had been to the seafood market in Wuhan, showed that the sequences were extremely similar to bat-derived SARS-like coronaviruses. Examining variable features such as its receptor-binding domain mutations and polybasic cleavage sites leads the authors to rule out a laboratory origin, and call for greater understanding of how zoonotic transmission occurs, to help prevent future events.
Genomics has played an outsized role in allowing scientists to develop therapeutics, diagnostics, and vaccines to combat COVID-19. Rapid COVID-19 tests, for instance, were developed thanks to the cloning and synthesis of specific SARS-CoV-2 viral proteins [25]. And over 70% of COVID-19 vaccine candidates were created through the use of genomics. Furthermore, like work in proteomics and metabolomics, genomics is capable of generating predictive models and identifying viral pathways capable of targeting. But unlike the former fields, genomic studies have introduced the element of ancestry into their associations regarding COVID-19 outcomes. Genomic studies identified a major genetic risk associated with COVID-19, spotlighting the association of a region of chromosome 3p21.31 with severe COVID-19—a gene of Neanderthal origin [26].
As such findings have emerged, the ethical implications around such knowledge have received some attention. Questions such as whether the genomic makeup of patients should influence vaccine allocation arose as it may recommend the prioritization of genetically vulnerable populations, while de-prioritizing socially marginalized groups of shared ancestry. The psychological aspects of genetic susceptibility being available have also been discussed in the literature, such as the possibility of undermining mass lockdown directives, should some groups be identified as less vulnerable than others. They also raise concerns about how genomic testing might threaten workplace privacy and discrimination, and recommend protections against such risks.
Genomics has also opened the door to improved transmission surveillance and patient monitoring (Fig. 2). Meredith (2020) integrated genomic and epidemiological data from almost 300 patients to identify viral clusters within an English hospital setting [27]. Using phylogenetic data to assess possible cases of transmission within wards allowed the researchers to inform the hospital management and infection control teams of possible reservoirs of patient infection, and to update their patient safety efforts and PPE procedures accordingly. On a national level, another study sequenced genomes from the second outbreak of COVID-19 in New Zealand during August 2020 [28]. As the sequences belonged to a single cluster, the scientists were able to determine that the second outbreak had been caused by a single introduction; and noted that close sharing of real-time viral genomic information with public health teams has allowed New Zealand to eliminate COVID-19 and prevent additional lockdowns. Similar genomic efforts have been put to use in Australia: where genomic clusters of infection within Victoria, Australia, demonstrated the role of returning travelers in driving domestic infection; and supported the effectiveness of efforts like quarantining such high-risk groups [29].
As with other omic fields, genomic researchers are now attempting to aggregate and share the results of their findings through open access platforms towards an effort to democratize data and advance science. The COVID-19 Host Genetics Initiative, launched in 2021, is an open-science collaboration that maps genetic determinants of COVID-19 infection in patients with respect to infection severity [30•]. The global network of genomic resources allowed researchers to identify 13 significant loci across the genome associated with COVID-19, with causal roles for smoking and BMI in severe COVID-19. Indeed, the creation of public viral genome data repositories changed the course of COVID-19 research: when the SARS-CoV-2 genome was first sequenced, the results were publicly deposited into GenBank, allowing the global research community to begin tackling the pandemic [31]. Open sharing of data has revolutionized research and our global efforts to direct these insights toward interventions.
From tracking and mapping to diagnosing and triaging, genomic data has enhanced our ability to understand, contain, and curb the virus. Knyazev refers to the “virtually unlimited potential” of rapidly generating and analyzing genomic data in our times [32]; the patient genomic data from Meredith was sequenced in under 24 h [33]. Perhaps the most tangible benefits have been genomics’ ability to identify contagion hotspots and disrupt transmission chains, such as those created from travel. Yet combining genomic data with epidemiological insights has allowed for researchers to make informed public health recommendations, and identify especially vulnerable groups who may require special assistance. In the event of a future pandemic, such tools will be indispensable.
COVID-Omics and Solid Organ Transplantation
The onset of the pandemic brought a dramatic reduction in transplantation across the world. While transplant activity stabilized after the first 3 months of the pandemic, the net loss of transplant opportunities for waiting list candidates amounted to about 50,000 years of waitlisted patient life-years lost [34]. As such, the unique health status and conditions of transplant patients require special protection against COVID-19. Impaired immune function from immunosuppressive medications or organ failure poses risks, and necessitates vaccination. Yet vaccination carries its own conditions: recipients of solid organ transplants have been shown to have a weak immune response to two doses of mRNA vaccine, and it is a third dose that significantly improves the immunogenicity of the vaccine [35]. Furthermore, despite reductions in surgical access due to COVID-19, lung transplantation has succeeded as a meaningful option for patients suffering from COVID-19-related respiratory failure [36].
The landscape of transplant multi-omics is home to an exciting and growing body of work–as is the intersection of transplant-omics and COVID-19 (Fig. 3). For instance, an early and highly cited paper by Wishart establishes the usefulness of metabolomic measurement as a diagnostic tool for donor organ viability, given the rapid metabolic responses to an “event,” relative to slower protein or tissue changes [37]. And Ramsey presents a broad, pandemic-oriented argument for the clinical utility of biomarker surveillance after solid organ transplant as a way of avoiding invasive biopsy procedures and limiting COVID-19 transmission by in-person exposure [38]. While the levels of uric metabolites and proteins have long been used by clinicians to gauge the health of a transplanted organ in kidney patients, omic data has been shown to better predict organ rejection compared to current benchmarks such as creatinine [39]. And whereas cycle threshold (CT) values are traditionally used by clinicians to predict severity and outcomes of disease, they have been found to be less useful for assessing COVID-19 severity in transplant patients [40]. Thus, COVID-19 omic studies have demonstrated great methodological diversity and creativity, from transcriptomic heat mapping of genes for viral gene expression [41]; to measuring concentrations of volatile organic compound (VOC) detection in patient’s exhaled breath for viral load [42•].
In renal COVID-omics, a noteworthy study is that of Rincon Arevalo, which recruited KTRs and dialysis patients to assess how their immune systems respond to mRNA vaccines relative to healthy populations [43]. Using single-cell transcriptome sequencing, the researchers found that immunosuppression in the KTRs and dialysis patients resulted in impaired post-vaccine immunity—supporting the call for triple vaccines, and urging scientists to ameliorate vaccination protocols for dialysis and kidney transplant patients. Another study by Sherwood combined immunological, serological, and proteomic monitoring to study a kidney transplant patient who developed symptomatic COVID-19 perioperatively. The researchers found that induction with basiliximab and maintenance of triple immunosuppression did not impair the patient’s ability to mount a strong viral immune responsible. And despite the limitations of a single case report, the authors’ findings suggest that clinicians may safely continue immunosuppression in patients with early post-transplant COVID-19 [44].
Sun is another critical study that seeks to explain the worsened COVID-19 outcomes experienced by KTRs relative to non-transplant patients [45]. The authors assess the transcriptomes of 64 COVID-positive KTRs, ranging from acute to post-acute conditions. The immune signature of acute disease was associated with decreased T-cell and adaptive immune activation despite reduction in immunosuppression, which then returned to normal in the post-acute phase, despite re-initiation of immunosuppression in the recovery phase. Their findings suggest that the current pandemic practice of reducing KTR immunosuppression might not prevent the COVID-induced impairment and subsequent recovery of adaptive immunity, and call for providers to reconsider their current therapeutic strategies.
Contrary to the above studies, a subsequent case study by Klein raised concern for immunosuppressive medication’s impact on renal transplant recipients in the setting of COVID-19. Using whole viral genomic sequencing to study a single kidney transplant patient who developed recurring COVID-19, the authors found that the reinfections were of distinct viral lineages, but the patient responded to the initial COVID-19 infection with a low neutralization titer. The authors suggested that immunosuppression lowered the patient’s CD4 T-cell pools and thus exposed the patient to reinfection [46].
Multi-omic research in transplantation has also unveiled new insights with regard to the lung transplant community. In a pioneering study of explanted lungs from the first successful lung transplant surgery on a COVID-19 patient in 2020, Chen used multi-omic analysis to reveal significant transcription and protein expression patterns consistent with fibrosis and inflammation. They concluded that significant neutrophil, T-cell, and macrophage activation leads to cytokine storm and fibrosis in the lungs, contributing to current understanding of COVID-19 pathogenesis [47]. Furthermore, the safety of lung transplantation from a COVID-positive lung donors has been a major safety concern, and led to reduced utilization of lungs from donors with recent COVID-19 diagnoses. Toward resolving this uncertainty, Saharia studied a transplant recipient hospitalized with hypoxemic respiratory failure following a positive COVID-19 test. Using genomic and subgenomic RNA analysis, it was found that genomic COVID-19 persisted in the donor lung tissue, but was not transmitted to the patient. Thus, the authors recommended that donor lungs containing COVID-19 genomic RNA can be safely transplanted with good outcomes [48].
While COVID-omics has thus contributed crucial insights in the nature and clinical management of COVID-19 in transplant patients, the body of research as a whole is still limited.
For instance, few omic studies have addressed the risks of COVID-19 in heart transplant patients. Lima suggests that COVID-19 negatively impacts heart transplantation patient outcomes, but is a retrospective analysis with an extremely small patient size [49]. Multi-omic research is also especially limited in liver transplantation, despite a broader movement in liver transplant studies toward precision medicine. Overall, this research shortage can be explained by the challenges and clinical priorities of the pandemic. Rincon-Avelo, Sun, and Kamp acknowledge their limited sample sizes; Sun attributes enrollment issues to hospital restrictions on in-person visits at the height of the pandemic. Furthermore, since the majority of COVID-omics work is humoral in focus, such transplant studies tend to emphasize immunity and organ viability but lack a deeper insight into the workings of the transplanted organ under the stress of COVID-19. Integrating plasma proteomics and organ proteomics may require multidimensional work along the lines of targeted sequencing for genomics, transcriptomics, and single-cell analysis, but prove rewarding in elucidating complex pathways and spotlighting opportunities for actionable interventions.
Conclusion
This review has surveyed the major contributions of multi-omic research to COVID-19 diagnostics and therapeutics, and evaluated the landscape of transplant COVID-omics. While specialized omic knowledge of transplant recipient health under COVID-19 has yet to be developed across the heart, kidney, liver, and lung, the robust body of COVID-omics knowledge indicates the potential of machine learning and high throughput technology to pinpoint and elucidate predictive biomarkers, patient risk stratification criteria, and biological pathways suitable for therapeutic intervention. Genomics has made particularly substantial COVID-19 public health contributions through patient tracking and cluster identification, allowing officials and hospital administration alike to reduce contact transmission.
The question of how COVID-omic insights might translate into clinical interventions, however, remains. The increasingly cheap cost of omic studies is generating more data, but whether or not it aids clinical decision-making is unclear. Clinical barriers to multi-omic applications may stem from a lack of data standardization and protocol transparency in the multi-omics communities. Increasing the participation of experienced clinicians in such omic studies might better guide and assess the output of such studies. Broader challenges also exist with the matter of integrating multi-omic insight into pharmaceutical pipelines. While unlocking multi-omic insights appears to be clearer than ever, the road to clinical and pharmaceutical translation remains slow and murky. Joint efforts between academic researchers, pharmaceutical companies, and public health organizations may be ideal in bridging these gaps.
Until these highlighted challenges can be resolved, the bedside impact of multi-omic research will be limited. However, the prospect of another pandemic—as well as the ongoing plights of long COVID-sufferers—proves the urgency of clarifying these matters. While COVID-omics might not have been rapid and coordinated enough to generate vaccine and pharmacological insights for this pandemic, omics remains a tremendously useful tool for non-time-sensitive cases. For instance, many individuals now live with long COVID, whose effects can be devastating, but the pathology itself remains little understood. For chronic diseases, the granular insight and wealth of data presented by multi-omics represents a tremendous opportunity to elucidate symptoms and mechanisms we still don’t understand. Furthermore, viruses as infective diseases cannot be underestimated in their power to harm the health of the world. Virologists are now calling for antiviral medicines that can be rapidly deployed, rather than using a “one-virus-one-drug approach.” If multi-omics can anticipate the next pandemic virus before it arrives, this, too, may be a life-saving use of time.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54.
Wu H, Soe MM, Konnor R, Dantes R, Haass K, Dudeck MA, et al. Hospital capacities and shortages of healthcare resources among US hospitals during the coronavirus disease 2019 (COVID-19) pandemic, National Healthcare Safety Network (NHSN), March 27-July 14, 2020. Infect Control Hosp Epidemiol. 2022;43(10):1473–6.
Zhuang Z, Cao P, Zhao S, Han L, He D, Yang L. The shortage of hospital beds for COVID-19 and non-COVID-19 patients during the lockdown of Wuhan. China. Ann Transl Med. 2021;9(3):200.
Acosta E. Global estimates of excess deaths from COVID-19. Nature. 2023;613(7942):31–3.
Yetkin Ozbuk RM, Coskun A, Filimonau V. The impact of COVID-19 on food management in households of an emerging economy. Socioecon Plann Sci. 2022;82:101094.
Pronk NP, McEvoy C. Equitable well-being, social trust, and the economy: An integrated health system's perspectives on the long-term implications of COVID-19. Prog Cardiovasc Dis. 2022;76:57–60.
Cisek K, Krochmal M, Klein J, Mischak H. The application of multi-omics and systems biology to identify therapeutic targets in chronic kidney disease. Nephrol Dial Transplant. 2016;31(12):2003–11.
Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. 2019;20(19):4781.
Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genomics. 2020;52(4):200–2.
Delafiori J, Navarro LC, Siciliano RF, de Melo GC, Busanello ENB, Nicolau JC, et al. Covid-19 Automated Diagnosis and Risk Assessment through Metabolomics and Machine Learning. Anal Chem. 2021;93(4):2471–9.
Zhou Z, Zhou X, Cheng L, Wen L, An T, Gao H, et al. Machine learning algorithms utilizing blood parameters enable early detection of immunethrombotic dysregulation in COVID-19. Clin Transl Med. 2021;11(9):e523.
• Overmyer KA, Shishkova E, Miller IJ, Balnis J, Bernstein MN, Peters-Clarke TM, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12(1):23–40 e7. Analyzes transcriptomic, proteomic, metabolomic, and lipidomic data to identify over 17,000 biomolecules in relation to COVID-19 patient outcomes, freely available on an interactive digital databae (covid-omics.app).
Nie X, Qian L, Sun R, Huang B, Dong X, Xiao Q, et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184(3):775-91 e14.
Wilk AJ, Lee MJ, Wei B, Parks B, Pi R, Martinez-Colon GJ, et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J Exp Med. 2021;218(8):e20210582.
• Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021;27(5):904–16. Uses single-cell transcriptomic analysis to provide a detailed illustration of the coordinated immune response to COVID-19 and identifies viable therapeutic targets.
Lancaster SM, Sanghi A, Wu S, Snyder MP. A Customizable Analysis Flow in Integrative Multi-Omics. Biomolecules. 2020;10(12):1606.
Thomas T, Stefanoni D, Reisz JA, Nemkov T, Bertolone L, Francis RO, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):e140327.
Paez-Franco JC, Maravillas-Montero JL, Mejia-Dominguez NR, Torres-Ruiz J, Tamez-Torres KM, Perez-Fragoso A, et al. Metabolomics analysis identifies glutamic acid and cystine imbalances in COVID-19 patients without comorbid conditions. Implications on redox homeostasis and COVID-19 pathophysiology. PLoS One. 2022;17(9):e0274910.
Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594(7862):246–52.
Suvarna K, Biswas D, Pai MGJ, Acharjee A, Bankar R, Palanivel V, et al. Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity With Drug Repurposing Potential. Front Physiol. 2021;12:652799.
Beaulieu-Jones BK, Yuan W, Brat GA, Beam AL, Weber G, Ruffin M, et al. Machine learning for patient risk stratification: standing on, or looking over, the shoulders of clinicians? NPJ Digit Med. 2021;4(1):62.
Callaway E, Cyranoski D, Mallapaty S, Stoye E, Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579(7800):482–3.
Larsen DA, Wigginton KR. Tracking COVID-19 with wastewater. Nat Biotechnol. 2020;38(10):1151–3.
Saravanan KA, Panigrahi M, Kumar H, Rajawat D, Nayak SS, Bhushan B, et al. Role of genomics in combating COVID-19 pandemic. Gene. 2022;823:146387.
Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600(7889):408–18.
Zeberg H, Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587(7835):610–2.
Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20(11):1263–72.
Douglas J, Geoghegan JL, Hadfield J, Bouckaert R, Storey M, Ren X, et al. Real-Time Genomics for Tracking Severe Acute Respiratory Syndrome Coronavirus 2 Border Incursions after Virus Elimination. New Zealand. Emerg Infect Dis. 2021;27(9):2361–8.
Seemann T, Lane CR, Sherry NL, Duchene S. Goncalves da Silva A, Caly L, et al. Tracking the COVID-19 pandemic in Australia using genomics. Nat Commun. 2020;11(1):4376.
• Initiative C-HG. Mapping the human genetic architecture of COVID-19. Nature. 2021;600(7889):472–7. Provides an overview of an open-science collaboration that maps COVID-19 genetic determinants in patients in relation to infection severity, resulting in the identification of closely-linked genomic loci and lifestyle behaviors.
Yang Y, Shang W, Rao X. Facing the COVID-19 outbreak: what should we know and what could we do? J Med Virol. 2020;92(6):536–7.
Knyazev S, Chhugani K, Sarwal V, Ayyala R, Singh H, Karthikeyan S, et al. Unlocking capacities of genomics for the COVID-19 response and future pandemics. Nat Methods. 2022;19(4):374–80.
Aubert O, Yoo D, Zielinski D, Cozzi E, Cardillo M, Durr M, et al. COVID-19 pandemic and worldwide organ transplantation: a population-based study. Lancet Public Health. 2021;6(10):e709–e19.
Kamar N, Abravanel F, Marion O, Couat C, Izopet J, Del Bello A. Three doses of an mRNA Covid-19 vaccine in solid-organ transplant recipients. N Engl J Med. 2021;385(7):661–2.
Roach A, Chikwe J, Catarino P, Rampolla R, Noble PW, Megna D, et al. Lung Transplantation for Covid-19-Related Respiratory Failure in the United States. N Engl J Med. 2022;386(12):1187–8.
Wishart DS. Metabolomics: the principles and potential applications to transplantation. Am J Transplant. 2005;5(12):2814–20.
Ramsey A. Genomic Biomarker Surveillance in the Care of Solid Organ Transplant Recipients: An Update for the General Clinician during the Coronavirus (CoVid-19) Pandemic. OBM Transplantation. 2020;4(2):1.
Lai X, Zheng X, Mathew JM, Gallon L, Leventhal JR, Zhang ZJ. Tackling chronic kidney transplant rejection: challenges and promises. Front Immunol. 2021;12:661643.
Gaston D, Malinis M, Osborn R, Peaper D, Landry M. Juthani-Mehta Manisha, Azar Marwan. Clinical implications of SARS-CoV-2 cycle threshold values in solid organ transplant recipients. Am J Transplant. 2021;21(3):1304–11.
Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–70.
Grassin-Delyle S, Roquencourt C, Moine P, Saffroy G, Carn S, Heming N, et al. Metabolomics of exhaled breath in critically ill COVID-19 patients: a pilot study. EBioMedicine. 2021;63:103154.
• Rincon-Arevalo H, Choi M, Stefanski AL, Halleck F, Weber U, Szelinski F, et al. Impaired humoral immunity to SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients and dialysis patients. Sci Immunol. 2021;6(60):eabj1031. Uses single-cell transcriptome sequencing to investigate immunosuppression in KTR and dialysis patients. Their findings of impaired post-vaccine immunity among such patients supported a call for triple vaccination among these populations, revising the existing standard of double vaccination.
Sun Z, Zhang Z, Banu K, Azzi YA, Reghuvaran A, Fredericks S, et al. Blood transcriptomes of SARS-CoV-2-infected kidney transplant recipients associated with immune insufficiency proportionate to severity. J Am Soc Nephrol. 2022;33(11):2108–22.
Sherwood K, Nicholl D, Fenninger F, Wu V, Wong P, Benedicto V, et al. Comprehensive immune profiling of a kidney transplant recipient with peri-operative SARS-CoV-2 infection: a case report. Front Immunol. 2021;12:753558.
Klein J, Brito A, Trubin P, Lu P, Wong P, Alpert T, et al. Longitudinal immune profiling of a severe acute respiratory syndrome coronavirus 2 reinfection in a solid organ transplant recipient. J Infect Dis. 2022;225(3):374–84.
Chen X, Lai K, Xu L, Yu Y, Wu B, He Y, Zhao W, et al. Novel insight from the first lung transplant of a COVID-19 patient. Eur J Clin Invest. 2021;51(1):e13443.
Saharia K, Ramelli S, Stein S, Roder A, Kreitman A, Banakis S. Successful lung transplantation using an allograft from a COVID-19–recovered donor: a potential role for subgenomic RNA to guide organ utilization. Am J Transplant. 2023;23(1):101–10.
Lima B, Gibson GT, Vullaganti S, Malhame K, Maybaum S, Hussain ST, et al. COVID-19 in recent heart transplant recipients: clinicopathologic features and early outcomes. Transpl Infect Dis. 2020;22(5):e13382.
Liu Z, Xu J, Que S, Geng L, Zhou L, Mardinoglu A, et al. Recent progress and future direction for the application of multiomics data in clinical liver transplantation. J Clin Transl Hepatol. 2022;10(2):363–73.
Acknowledgements
The authors are grateful to the patients whose lives and contributions made possible the discoveries and interventions discussed in this review. We also thank Daniel Viray and BioRender for creating the figures seen in this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have no conflicts of interest.
Human and Animal Rights
As this was a review, human and animal rights as well as informed consent were per each study included and do not apply to the work of the current authors. Same with ethics guidelines. We are happy to include some language around this if the editors think it should be still included in acknowledgments.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, A.A., Mansour, S.G. Improving the Odds—COVID-Omics and Predicting Patient Outcomes. Curr Transpl Rep 10, 126–134 (2023). https://doi.org/10.1007/s40472-023-00403-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40472-023-00403-7