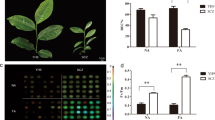
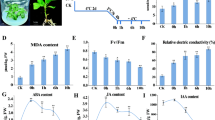
Abstract
Low temperatures limit crop growth, yield, and quality in cold areas, and cold tolerance is the only strategy for plants to survive in those areas. However, mulberry’s (Morus alba L.) molecular regulatory mechanisms in response to cold remain poorly understood. In this report, four cDNA libraries were built from two groups, the cold-treated group and the control group, and evaluated by RNA-Seq analysis. A total of 3593 differential expression genes (DEGs) were identified from 20,279 unigenes. Among these, 2337 up-regulated and 1256 down-regulated DEGs were identified under this standard (corrected p value < 0.05, log2fold change > 0.0). Most of these DEGs are involved in plant hormone signal transduction, MAPK signaling pathway, ubiquinone, and other terpenoid-quinone biosynthesis, nitrogen metabolism, and biosynthesis of secondary metabolites. A series of candidate genes related to cold stress were screened out and discussed based on these results. Furthermore, the expression changes of 8 genes under cold stress from RNA-Seq analysis were further validated by real-time quantitative polymerase chain reaction. This research reflects Morus alba L. transcriptome data and may help elucidate the cold response mechanisms of mulberry trees and would be useful in breeding cold-tolerant mulberry plants.



Similar content being viewed by others
Abbreviations
- DEGs:
-
Differential expression genes
- cDNA:
-
Complementary deoxyribonucleic acid
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- PCR:
-
Polymerase chain reaction
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Aoki-Kinoshita KF, Kanehisa M (2007) Gene annotation and pathway mapping in KEGG. In: Comparative genomics. Springer, pp 71–91
Ariyadasa R, Mascher M, Nussbaumer T, Schulte D, Frenkel Z, Poursarebani N, Zhou R, Steuernagel B, Gundlach H, Taudien S, Felder M (2014) A sequence-ready physical map of barley anchored genetically by two million single-nucleotide polymorphisms. Plant Physiol 164:412–423
Bai X, Long J, Li S, Li K, Xu H (2014) Molecular cloning and characterisation of a germin-like protein gene in spinach (SoGLP). J Hortic Sci Biotechnol 89:592–598
Bakshi M, Oelmüller R (2014) WRKY transcription factors: jack of many trades in plants. Plant Signal Behav 9:27700
Beeckman T, Przemeck GK, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130:1883–1893
Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29:23–32
Boller T (1988) Ethylene and the regulation of antifungal hydrolases in plants. Oxford Surv Plant Mol Cell Biol 5:145–174
Bosch M, Hepler PK (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17:3219–3226
Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17:2281–2295
Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52:1154–1168
Chandler JW (2018) Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci 23:151–162
Chauhan S, Devi U, Kumar VR, Kumar V, Anwar F, Kaithwas G (2015) Dual inhibition of arachidonic acid pathway by mulberry leaf extract. Inflammopharmacology 23:65–70
Chen L, Yang Y, Liu C, Zheng Y, Xu M, Wu N, Sheng J, Shen L (2015) Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. BBRC 464:962–968
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582
Cheng XX, Zhao LH, Klosterman SJ, Feng HJ, Feng ZL, Wei F, Shi YQ, Li ZF, Zhu HQ (2017) The endochitinase VDECH from Verticillium dahliae inhibits spore germination and activates plant defence responses. Plant Sci 259:12–23
Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224:1174–1184
Choi J, Kang HJ, Kim SZ, Kwon TO, Jeong SI, Jang SI (2013) Antioxidant effect of astragalin isolated from the leaves of Morus alba L. against free radical-induced oxidative hemolysis of human red blood cells. Arch Pharm Res 36:912–917
Dong C-J, Liu J-Y (2010) The Arabidopsis EAR-motif-containing protein RAP2. 1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol 10:47
Du C, Hu K, Xian S, Liu C, Fan J, Tu J, Fu T (2016) Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus L.). Mol Genet Genomics 291:1053–1067
Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signalling to jasmonate and ethylene responses. Plant Cell 14:1557–1566
Fukaki H, Fujisawa H, Tasaka M (1996) Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol 110:933–943
Gomat HY, Deleporte P, Moukini R, Mialounguila G, Ognouabi N, Saya AR, Vigneron P, Saint-Andre L (2011) What factors influence the stem taper of Eucalyptus: growth, environmental conditions, or genetics? Ann For Sci 68:109–120
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hincha DK, Meins F Jr, Schmitt JM (1997) [beta]-1, 3-Glucanase is cryoprotective in vitro and is accumulated in leaves during cold acclimation. Plant Physiol 114:1077–1083
Hu Xiaoqing, Xu Xuemei, Li Chenghao (2018) Ectopic expression of the LoERF017 transcription factor from Larix olgensis enhances salt and osmotic-stress tolerance in Arabidopsis thaliana. Plant Biotechnol Rep 12:93–104
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. MGG 244:563–571
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276:3148–3162
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Juge N (2006) Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci 11:359–367
Keller F, Pharr DM (2017) Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides. In: Photoassimilate distribution plants and crops source-sink relationships, Routledge, pp 157–183
Kosuge T, Nester EW (1984) Plant-microbe interactions; molecular and genetic perspectives. Oxford University Press (OUP) 36(5):343–344
Li B, Dewey C (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 12:323
Li Y, Qian Q, Zhou Y, Ya M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li S, Bashline L, Lei L, Gu Y (2014) Cellulose synthesis and its regulation. In: The Arabidopsis Book/American Society of Plant Biologists, vol 12, p e0169
Li P, Liu Y, Tan W, Chen J, Zhu M, Lv Y, Liu Y, Yu S, Zhang W, Cai H (2019) Brittle Culm 1 encodes a COBRA-like protein involved in secondary cell wall cellulose biosynthesis in Sorghum. Plant Cell Physiol 60:788–801
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu J, Shi Y, Yang S (2018) Insights into the regulation of C-repeat binding factors in plant cold signaling. J Integr Plant Biol 60:780–795
Lu M, Han Y-P, Gao J-G, Wang X-J, Li W-B (2010) Identification and analysis of the germin-like gene family in soybean. BMC Genom 11:620
Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S (2006) Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol 141:47–60
Ma L, Coulter JA, Liu L, Zhao Y, Chang Y, Pu Y, Zeng X, Xu Y, Wu J, Fang Y, Bai J (2019) Transcriptome analysis reveals key cold-stress-responsive genes in winter rapeseed (Brassica rapa L.). Inter J Mol Sci 20:1071
McCann M, Rose J (2010) Blueprints for building plant cell walls. Plant Physiol 152(2):365–365
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Miyashita Y (1986) A report on mulberry cultivation and training methods suitable for bivoltine rearing in Karnataka. Central Silk Board, Bangalore, pp 1–7
Morozova O, Hirst M, Marra MA (2009) Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genom Hum Genet 10:135–151
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621
Müller K, Levesque-Tremblay G, Bartels S, Weitbrecht K, Wormit A, Usadel B, Haughn G, Kermode AR (2013) Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol 161:305–316
Nakagawa K (2013) Studies targeting α-glucosidase inhibition, antiangiogenic effects, and lipid modification regulation: background, evaluation, and challenges in the development of food ingredients for therapeutic purposes. Biosci Biotechnol Biochem 77:900
Naowaboot J, Pannangpetch P, Kukongviriyapan V, Kongyingyoes B (2009) Antihyperglycemic, antioxidant and antiglycation activities of mulberry leaf extract in streptozotocin-induced chronic diabetic rats. Plant Food Hum Nutr 64:116–121
Neven LG, Haskell DW, Guy CL, Denslow N, Klein PA, Green LG, Silverman A (1992) Association of 70-kilodalton heat-shock cognate proteins with acclimation to cold. Plant Physiol 99:1362–1369
Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1, 4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17:5563–5576
Pagter M, Alpers J, Erban A, Kopka J, Zuther E, Hincha DK (2017) Rapid transcriptional and metabolic regulation of the deacclimation process in cold acclimated Arabidopsis thaliana. BMC Genom 18:731
Peng X, Wu Q, Teng L, Tang F, Pi Z, Shen S (2015) Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol 15:108
Pennycooke JC, Cox S, Stushnoff C (2005) Relationship of cold acclimation, total phenolic content and antioxidant capacity with chilling tolerance in petunia (Petunia hybrida). Environ Exp Bot 53:225–232
Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. PNAS 102:8633–8638
Phan TD, Bo W, West G, Lycett GW, Tucker GA (2007) Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol 144:1960–1967
Phukan S, Ujjal J, Gajendra SJ, Rakesh K (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7:760
Rahmathulla VK (2012) Management of climatic factors for successful silkworm (Bombyx mori L.) crop and higher silk production: a review. Psyche J Entomol 2012:1–12
Rao MV, Lee HI, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12:1633–1646
Riechmann JL, Heard J, Martin M, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Eur PMC 290:2105–2110
Rohela GK, Phanikanth J, Aftab AS, Pawan S, Sadanandam A, Mrinal K (2018) In vitro regeneration and assessment of genetic fidelity of acclimated plantlets by using ISSR markers in PPR-1 (Morus sp.): an economically important plant. Sci Hortic 241:313–321
Saibo NJ, Lourenço T, Oliveira MM (2009) Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot 103:609–623
Scheible WR, Pauly M (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7:285–295
Schluttenhofer C, Ling Y (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 167:295–306
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C T method. Nat Protoc 3:1101
Shimosaka E, Ozawa K (2015) Overexpression of cold-inducible wheat galactinol synthase confers tolerance to chilling stress in transgenic rice. Breed Sci 65:363–371
Shukla P, Gulab KR, Aftab AS, Sharma SP (2016) Prospect of Cold Tolerant Genes and Its Utilization in Mulberry Improvement. Ind Hortic J 6:127–129
Simon SA, Zhai J, Nandety RS, McCormick KP, Zeng J, Mejia D, Meyers BC (2009) Short-read sequencing technologies for transcriptional analyses. Annu Rev Plant Biol 60:305–333
Singh VK, Jain M (2015) Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front Plant Sci 6:918
Soga K, Kotake T, Wakabayashi K, Kamisaka S, Hoson T (2008) Transient increase in the transcript levels of γ-tubulin complex genes during reorientation of cortical microtubules by gravity in azuki bean (Vigna angularis) epicotyls. J Plant Res 121:493–498
Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S (2004) Toward a systems approach to understanding plant cell walls. Science 306:2206–2211
Song Y, Wang L, Xiong L (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229:577–591
Song J, Xing Y, Munir S, Yu C, Song L, Li H, Ye Z (2016) An ATL78-Like RING-H2 finger protein confers abiotic stress tolerance through interacting with RAV2 and CSN5B in tomato. Front Plant Sci 7:1305
Sugiyama M, Katsube T, Koyama A, Itamura H (2016) Effect of solar radiation on the functional components of mulberry (Morus alba L.) leaves. J Sci Food Agri 96:3915–3921
Sun Q, Wang G, Zhang X, Zhang X, Qiao P, Long L, Yuan Y, Cai Y (2017) Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci Rep 7:1–9
Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126:45–51
Tang M, Liu X, Deng H, Shen S (2011) Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Sci 181:623–631
Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12:2529–2539
Tian M, Lou L, Liu L, Yu F, Zhao Q, Zhang H, Zhu B (2015) The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J 82:81–92
Torres LF, Reichel T, Déchamp E, de Aquino SO, Duarte KE, Alves GSC et al (2019) Expression of DREB-like genes in Coffea canephora and C. arabica subjected to various types of abiotic stress. Trop Plant Biol 12:98–116
Trujillo LE, Sotolongo M, Menendez C, Ochogavia ME, Coll Y, Hernandez I, Borras-Hidalgo O, Thomma BP, Vera P, Hernandez L (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49:512–525
Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, Guilfoyle TJ, Chen M, Qi Y (2010) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genom 10:533–546
Wang H, Zhou L, Fu Y, Cheung MY, Wong FL, Phang TH, Lam HM (2012) Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant Cell Environ 35:1932–1947
Wang H, Tong W, Feng L, Jiao Q, Long L, Fang R, Zhao W (2014) De novo transcriptome analysis of mulberry (Morus L.) under drought stress using RNA-Seq technology. Russ J Bioorg Chem 40:423–432
Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63:381–407
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10:228
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. MGG 238:17–25
Yang QS, Wu JH, Li CY, Wei YR, Sheng O, Hu CH, Kuang RB, Huang YH, Peng XX, McCardle JA, Chen W (2012) Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB Group) seedlings. Mol Cell Proteom 11:1853–1869
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Zhang H, Liu W, Wan L, Li F, Dai L, Li D, Zhang Z, Huang R (2010) Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Trans Res 19:809–818
Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y (2012) De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.). BMC Genom 13:90
Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X (2013) Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 8:10
Zhang Y, Yu H, Yang X, Li Q, Ling J, Wang H, Gu X, Huang S, Jiang W (2016) CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol Biochem 108:478–487
Zhao Z, Tan L, Dang C, Zhang H, Wu Q, An L (2012) Deep-sequencing transcriptome analysis of chilling tolerance mechanisms of a subnival alpine plant, Chorispora bungeana. BMC Plant Biol 12:222
Acknowledgements
This work was supported by Sericulture Industry Technology in China Agriculture Research System (CARS-18-ZJ0207), Guangxi innovation driven development project (AA19182012-2), Open Program of Key Laboratory of Silkworm and Mulberry Genetic Improvement Ministry of Agriculture, China (KL201906), the Crop Germplasm Resources Protection Project of the Agriculture Ministry (111721301354052026), and National Infrastructure for Crop Germplasm Resources (NICGR-43).
Author information
Authors and Affiliations
Contributions
AA, LL, LQ, and ZW conceived and designed research. AA, LY, and DQ conducted experiments. MA, ZD, and GP contributed new reagents or analytical tools. AA, SY, and QC analyzed data. AA and LL wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adolf, A., Liu, L., Ackah, M. et al. Transcriptome profiling reveals candidate genes associated with cold stress in mulberry. Braz. J. Bot 44, 125–137 (2021). https://doi.org/10.1007/s40415-020-00680-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-020-00680-x