Abstract
Background
Mycoplasma pneumoniae (MP) is the most common pathogen of atypical pneumonia and the main cause of community-acquired pneumonia (CAP) in infants and older adults. This study aimed at investigating a method based on the cross-priming amplification (CPA) technique for the rapid detection of MP in clinical specimens collected from patients with CAP.
Methods
The sensitivity and specificity of the EasyNAT MP assay were determined. Oropharyngeal swab specimens were collected from 162 in-patients of Hangzhou First People’s Hospitals from January 2018 to December 2020. The patients were aged between 1 and 15 years with symptoms, signs, and chest radiographs consistent with CAP. This study evaluated the presence of MP in the clinical specimens using the EasyNAT method and the conventional fluorescence quantitative PCR technique.
Results
The limit of detection using the EasyNAT MP assay was 500 copies/mL, while the test results of the other 13 common pathogens causing CAP or colonizing in the upper respiratory tract showed no cross-reactivity. Of 162 specimens, EasyNAT MP gave a positive indication in 82 specimens. Compared with conventional fluorescence quantitative PCR, the positive coincidence rate and the negative coincidence rate of EasyNAT MP was found to be 100.00% and 97.56%, respectively. Of the 82 specimens, two specimens were determined to be negative by the conventional fluorescence quantitative PCR, but were positive for EasyNAT MP. The two samples were re-extracted and confirmed to be positive by conventional fluorescence quantitative PCR.
Conclusion
EasyNAT MP is suitable as an initial test for MP diagnosis due to its simplicity, low turnaround time, and high sensitivity and specificity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Mycoplasma pneumoniae (MP) is one of the most common pathogens causing community-acquired pneumonia. |
The culture and serological tests commonly used in the laboratory are not suitable for the routine detection of MP infection. Due to the need for technical expertise and sophisticated laboratory equipment, the application of conventional fluorescence quantitative PCR in the diagnosis of MP infection is also limited. |
The EasyNAT MP assay is suitable for use as an initial test for MP diagnosis due to its simplicity, low turnaround time, and high sensitivity and specificity. |
1 Introduction
Mycoplasma pneumoniae, the most common pathogen of atypical pneumonia, mainly causes community-acquired pneumonia (CAP) in infants and older adults, especially between the ages of 6 and 10 years [1] and between the ages of 25 and 44 years [2]. M. pneumoniae pneumonia (MPP) accounts for 4–8% of the etiology of community-acquired bacterial pneumonia, up to 20% during the epidemic seasons and 70% in close-contact crowds [3,4,5]. Although most MPPs are mild or self-limiting, a small number of MPPs can still lead to severe symptoms and death. In addition, M. pneumoniae infection can lead to severe extrapulmonary diseases including M. pneumoniae-induced rash and mucositis (MIRM) [6], erythema multiforme (EM) [7], and Stevens–Johnson syndrome-toxic epidermal necrosis (SJS-TEN) [8], as well as neurological and hematological symptoms. It can also cause arthritis in patients with primary humoral immunodeficiency [9] and asthma in children [10]. Unlike other bacterial pathogens including Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis, β-lactam antibiotics used for empirical therapy are ineffective against M. pneumoniae. Thus early, rapid and effective detection of the pathogen can be useful for commencement of appropriate antimicrobial therapy.
Laboratory methods for detecting M. pneumoniae include culture, serology, and nucleic acid amplification techniques. M. pneumoniae culture testing is not routinely carried out in the clinical laboratory because M. pneumoniae grows slowly and takes 3–4 weeks or more to form visible clones [11]. The antibody titer is increased fourfold or more in the acute phase in comparison to the convalescent phase, and is recognized as the gold standard for serological diagnosis of infectious diseases, thus only allowing for a retrospective diagnosis [11]. Target genomic DNA detection methods, such as fluorescence quantitative PCR (qPCR), provide a fast and sensitive method to detect pathogens. However, technical expertise and sophisticated laboratory equipment are required, which are not always readily available, particularly in primary hospitals [11]. M. pneumoniae can be detected by liquid culture, as its decomposition of glucose produces acid, which can change the liquid medium pH and lead to the discoloration of phenol red indicator. As M. salivarium, M. orale, and others are part of the normal flora population in the upper respiratory tract, it is often difficult to interpret the results. To date, the US Food and Drug Administration (FDA) has not approved a point-of-care testing (POCT) reagent for rapid detection of M. pneumoniae.
The assay is based on a promising and innovative nucleic acid amplification technique called cross-priming amplification (CPA), which is highly efficient in providing rapid results. Rapid amplification of the target sequence is performed using isothermal conditions (55–65 °C), which obviates the requirement for technical expertise and complicated laboratory equipment such as a thermocycler [12]. CPA technology has been applied in the detection of Mycobacterium tuberculosis [13], and some studies have been performed to detect Pediococcus acidilactici and Babesia motasi [14, 15]. Another isothermal amplification technique, which is called loop-mediated isothermal amplification (LAMP), is widely favored at the beginning of development because of isothermal amplification, and the products were shown by direct visualization of a color change after staining [16]. DNA extraction is needed and further analysis of the product requires the help of the other PCR techniques, such as nucleic acid purification, gel electrophoresis, and fluorescence PCR [17]. These shortcomings limit the wide use of LAMP in clinical laboratories.
This study assessed the sensitivity and specificity of the CPA technique in comparison to the qPCR method. To our knowledge, there is a paucity of data concerning the detection of M. pneumoniae via the CPA method in China, prompting us to investigate a CPA technique for the rapid detection of M. pneumoniae in clinical specimens in Hangzhou, China.
2 Materials and Methods
2.1 The Mycoplasma pneumoniae Cartridge Used in this Study
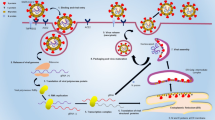
The EasyNAT MP (Ustar Biotechnologies Ltd, Hangzhou, China) assay has multiple hydrophobic separation layers to isolate the lysate, the cleaning solution, and the reaction solution in the cartridge (Fig. 1). In order to test the sample, the extract is chemically cracked at 95 °C to release the nucleic acid in the nucleic acid analyzer (Ustar Biotechnologies Ltd). The nucleic acid of the specimen used for detection passes through different liquid layers by means of magnetic permeability of the nucleic acid analyzer, and finally the nucleic acid is eluted in the cartridge leg and an expansion reaction occurs. The target sequence of MP DNA can be amplified isothermally with amplification primers, accelerated primers and cross primers using Bst DNA polymerase. MP can be detected by fluorescence labeled probe during the amplification process by the nucleic acid analyzer, and the results can be reported directly. This is a "one-tube" fully-automated nucleic acid analysis, that is, the cleavage binding, cleaning, elution, and amplification reaction are completed in the closed cartridge.
2.2 Primers and Probe Used in EasyNAT
Referring to the standard generic reference strain sequence of M. pneumoniae (NCTC 10119, LR214945.1), primers and probe were designed for its highly conserved p1 gene (Table 1 and Fig. 2). Primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
2.3 Generation of a Standard DNA Template
Aimed at generating a M. pneumoniae standard DNA for the CPA technique, a 192 bp PCR product from the p1 gene was amplified from the M. pneumoniae DNA using 5´-GGATCC (BamHI) GTGAACGTATCGTAACACGAGCTT-3´ and 5´-GTCGAC (SalI) TCATACCGGCGTAACGCAAAG-3´ as primers, and then cloned into the plasmid PUC57(1) (Takara, Dalian, China) to produce standard templates. The generated plasmid, PUC57(1)p1, was transformed into Escherichia coli DH5α (Takara, Dalian, China) cells and the positive clones were identified by sequencing (Sangon Biotech, Shanghai, China) with the primers listed above. PUC57(1)p1 was purified with the SanPrep Plasmid MiniPrep Kit (Sangon Biotech, Shanghai, China) and quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Shanghai, China). The copy number of DNA molecules was calculated according to the following formula: amount (copies/μl) = [DNA concentration (g/μl)/(plasmid length in base pairs × 660)] × 6.02 × 1023. Aliquots of the standard DNA were prepared in tenfold serial dilutions from 5.0 × 103 to 5.0 × 102 copies / mL in nuclease-free water and stored at − 80 °C until used.
2.4 Sensitivity and Specificity Assessment of EasyNAT
The sensitivity of the EasyNAT for M. pneumoniae detection was studied using plasmid containing target fragment, which was constructed as described above. An evaluation of the concentration and quality of the extracted DNA was then assessed by the NanoDrop 2000 absorption spectra. The plasmid underwent serial dilution (0.08 M EDTA, 0.1 M Tris) to obtain 5000, 1000, 500, and 100 copies/ml. Then 500 μl plasmid samples were added for detection. The detection results were interpreted automatically by the nucleic acid analyzer. Each diluted pathogen was tested 20 times, and the lowest dilution concentration that was positive in all 20 replicates defined as the limit of detection.
In order to investigate the specificity of the detection system, extracted DNA from the following pathogens were used as templates: E. coli, Staphylococcus aureus, Klebsiella pneumoniae, Bordetella pertussis, Legionella pneumophila, Streptococcus pneumoniae, Haemophilus influenzae, Ureaplasma urealyticum, Chlamydia pneumoniae, M. salivarium, M. orale, and Influenza A and B viruses, which were clinical isolates or nucleic acid-positive samples confirmed by sequencing. K. pneumoniae was available from the Microbiology Laboratory of Hangzhou First People's Hospital, C. pneumoniae, M. salivarium and M. orale were donated by Weiyuan Gene Technology Co., Ltd (Guangzhou), and other pathogens were available from Ustar Biotechnologies Ltd (Hangzhou). The specificity of the detection system was performed by EasyNAT MP on 0.5 Mcfarland of each pathogen. The detection results were interpreted automatically by the nucleic acid analyzer. Each pathogen was tested three times and the results that were negative in all three replicates were considered to be accurate.
2.5 Clinical Application of M. pneumoniae Detection
Previously healthy children aged between 1 and 15 years with symptoms, signs, and chest radiographs consistent with CAP who had been examined in the Pediatrics Department of Hangzhou First People's Hospital in Zhejiang, China from January 2018 to December 2020 were prospectively enrolled in this study. The oropharyngeal swab samples collected in this study were stored by the Gene Laboratory of Hangzhou First People's Hospital. The oropharyngeal swab samples that had been collected were washed with 3.0 ml sterile saline, and the washing solution was divided into two parts on average. This study has been licensed by the Hangzhou First People's Hospital Medical Technology Clinical Application and Scientific Research Ethics Committee (KY-20211203-0127-01).
2.5.1 Fluorescent qPCR
One part of the collected oropharyngeal swab samples was tested with fluorescence quantitative PCR as a control. The sample of the eluent was centrifuged at 12,000 revolutions per minute (rpm) for 5 min. The supernatant was discarded and the precipitate mixed with 1 ml sterilized saline and centrifuged at 12,000 rpm for 5 min. Then 50 μl lysate (1 M Tris-HCL pH 8.0, 1 mmol/L EDTA, 1% TritonX-100) was added to the precipitate and mixed well. The eluent was boiled for 10 min at 100 ℃ constant temperature and then centrifuged at 12,000 rpm for 5 min. The supernatant was then ready for using. An M. pneumoniae nucleic acid detection kit (DaAn Gene Co., Ltd. Guangzhou) was used with a ABI7500 (Thermo Fisher Scientific, USA) fluorescence quantitative PCR instrument and the operational steps were performed in strict accordance with the kit instructions.
2.5.2 EasyNAT Assay
The other half of the collected oropharyngeal swab sample, which had been added to the MP-DNA extraction solution after having been well mixed, was added to the MP cartridge. The two-dimensional code of the detection tube faces the scanning area of the nucleic acid analyzer and is the interface by which the information from the detection tube is analyzed. The detection tube was put into the sample chamber of the nucleic acid analyzer. The instrument begins to run the test after closing the lid of the sample chamber and clicking the start button. The instrument automatically displays and saves the test results when the cycles are finished (Fig. 3). The experiment was repeated once with the samples with results not consistent with those of the control fluorescence quantitative PCR.
2.6 Statistical Analyses
The differences between the detection rates of the two methods were tested using McNemar’s test. A P value of < 0.01 was considered statistically significant.
3 Results
3.1 Sensitivity and Specificity of EasyNAT for M. pneumoniae Detection
Analysis of the 13 pathogens detected showed no positive results, indicating that the specificity of the EasyNAT MP assay was good. The detection limit of plasmid PUC57(1)p1 was 500 copies/mL (Tables 2 and 3).
3.2 Clinical Performance of EasyNAT M. pneumoniae Assay
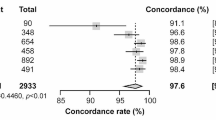
A total of 162 clinical samples were analyzed by the EasyNAT MP assay, and the results showed that 82 cases were positive and 80 cases negative. Compared with the control DaAn Gene fluorescence quantitative PCR, the positive coincidence rate was 100%, the negative coincidence rate was 97.56%, and the total coincidence rate 98.77%. The results are shown in Table 4.
3.3 Comparison of EasyNAT MP Assay with the Conventional PCR/RT-PCR Method
Compared with the traditional application method, the newly established isothermal amplification technique based on CPA is rapid, simple, and suitable for bedside detection of M. pneumoniae. The comparison is shown in Table 5.
4 Discussion
Although culture and immunohistochemical detection of original samples are the gold standard for diagnosis of infectious diseases, it is extremely difficult to accurately evaluate their performance due to the particularity of M. pneumonia [18]. Despite the lack of extensive culture verification, the higher analytical sensitivity and shorter turnaround time of nucleic acid amplification tests (NAATs) make it the "new gold standard" to detect pathogens of infectious disease. NAAT protocols require the ability to amplify a unique sequence region of the DNA from the infectious organism. The most widely accepted method for NAATs is fluorescent qPCR. The requirement of instrumentation, the complexity of procedures, and the associated costs decrease the usefulness of qPCR in primary hospitals. While a number of isothermal nucleic acid amplification methods have been developed to overcome these issues, they involve relatively simple protocols requiring the use of multiple enzymes and special primers/probe.
As a one-step diagnostic system, EasyNAT MP can detect MP in clinical samples and show 100% specificity for several pathogens that cause CAP. The CPA-NAATS system is based on isothermal DNA amplification technology, which is easy to operate and does not need additional equipment. The amplification of M. tuberculosis DNA by double-crossing CPA has been applied to the clinical diagnosis of tuberculosis patients [12]. In this study, a single-crossing CPA assay is used, which is slightly different to a double-crossing CPA assay [19]. We have successfully adopted this technology and optimized all the reaction conditions to achieve the purpose of developing a MP detection system. Compared with the conventional fluorescence quantitative PCR, the total coincidence rate of the EasyNAT MP is 98.77%.
Although the loop-mediated isothermal amplification (LAMP) has provided highly efficient results in detecting M. pneumoniae in clinical specimens of patients with CAP [17], it is not suitable for clinical POCT detection because of the requirement to analyze PCR amplification products. CPA technology is a powerful innovative nucleic acid isothermal amplification system developed by Ustar Biotechnologies (Hangzhou) Ltd [19]. The target sequence of DNA can be amplified isothermally with amplification primers, accelerated primers, and cross primers using Bst DNA polymerase, which has high activity to accomplish the chain replacement at 63 ℃. Similar to Xpert MTB/RIF (Cepheid, USA), EasyNAT MP uses preloaded reagents in a single cartridge that accommodates DNA extraction, DNA purification, and target sequence amplification and detection using three separate chambers within the same cartridge [11]. The main focus of EasyNAT is the design of specific CPA primers and the optimization of concentration and the proportion of primers and probes so that M. pneumoniae can be identified rapidly and sensitively.
In this study, we evaluated the CPA isothermal amplification with the EasyNAT MP assay to determine whether it could accurately detect M. pneumoniae in clinical oropharyngeal swab specimens. EasyNAT MP is more sensitive and reliable than the conventional fluorescence quantitative PCR technique for the detection of M. pneumoniae. In general, the EasyNAT MP assay’s simplicity of use, convenience, cheapness, and suitability for POCT are significantly advantageous when compared to traditional technology. It has good consistency and can meet the needs of clinical diagnosis. However, like other PCR technologies, the EasyNAT MP results depend on the quality of the collected samples. In addition, the isothermal amplification technology used in a CPA-NAATS system is non-exponential amplification. The cycle threshold (CT) is lack of quantitative relationship with the number of DNA templates, so EasyNAT MP detection is a qualitative test rather than a quantitative test.
5 Conclusion
The EasyNAT MP assay is suitable for use as an initial test for MP diagnosis due to its simplicity, low turnaround time, and high sensitivity and specificity.
References
Gao LW, Yin J, Hu YH, Liu XY, Feng XL, He JX, Liu J, Guo Y, Xu BP, Shen KL. The epidemiology of paediatric Mycoplasma pneumoniae pneumonia in North China: 2006 to 2016. Epidemiol Infect. 2019;147:e192.
Beeton ML, Zhang XS, Uldum SA, Bébéar C, Dumke R, Gullsby K, Ieven M, Loens K, Nir-Paz R, Pereyre S, Spiller OB, Chalker VJ. Mycoplasma pneumoniae infections, 11 countries in Europe and Israel, 2011 to 2016. Euro Surveill. 2020;25(2):pii=1900112.
Kong DC, Wu HY, Zheng YX, Pan H, Jiang CY, Zhang X, Chen J, Wu F. Etiologic and epidemiologic features of acute respiratory infections in adults from Shanghai, during 2015–2017. Zhonghua liu xing bing xue za zhi Zhonghua liuxingbingxue zazhi. 2019;40(8):904–10.
Lee E, Kim CH, Lee YJ, Kim HB, Kim BS, Kim HY, Kim Y, Kim S, Park C, Seo JH, et al. Annual and seasonal patterns in etiologies of pediatric community-acquired pneumonia due to respiratory viruses and Mycoplasma pneumoniae requiring hospitalization in South Korea. BMC Infect Dis. 2020;20(1):132.
Dierig A, Hirsch HH, Decker ML, Bielicki JA, Heininger U, Ritz N. Mycoplasma pneumoniae detection in children with respiratory tract infections and influence on management—a retrospective cohort study in Switzerland. Acta Paediatr 2020;109(2):375–80.
Zao I, Ribeiro F, Rocha V, Neto P, Matias C, Jesus G. Mycoplasma pneumoniae-associated Mucositis: a recently described entity. Eur J Case Rep Intern Med. 2018;5(11):000977.
Amode R, Ingen-Housz-Oro S, Ortonne N, Bounfour T, Pereyre S, Schlemmer F, Bequignon E, Royer G, Wolkenstein P, Chosidow O. Clinical and histologic features of Mycoplasma pneumoniae-related erythema multiforme: a single-center series of 33 cases compared with 100 cases induced by other causes. J Am Acad Dermatol. 2018;79(1):110–7.
Watkins LKF, Olson D, Diaz MH, Lin X, Demirjian A, Benitez AJ, Winchell JM, Robinson CC, Bol KA, Glodé MP, et al. Epidemiology and molecular characteristics of mycoplasma pneumoniae during an outbreak of M. pneumoniae-associated Stevens-Johnson Syndrome. Pediatric Infect Dis J. 2017;36(6):564–71.
de Groot RCA, Meyer Sauteur PM, Unger WWJ, van Rossum AMC. Things that could be Mycoplasma pneumoniae. J Infect. 2017;74(Suppl 1):S95-s100.
Kumar S, Roy RD, Sethi GR, Saigal SR. Mycoplasma pneumoniae infection and asthma in children. Trop Doct. 2019;49(2):117–9.
Dien Bard J, Hong T, Prichard M, Brooks E, Dallas S, Duffy L, Mixon E, Fowler KB, Atkinson TP, Parrott GL, et al. A Compendium for Mycoplasma pneumoniae. J Clin Microbiol. 2016;7:513.
Zhang Z, Du J, Liu T, Wang F, Jia J, Dong L, Zhao L, Xue Y, Jiang G, Yu X, et al. EasyNAT MTC assay: a simple, rapid, and low-cost cross-priming amplification method for the detection of mycobacterium tuberculosis suitable for point-of-care testing. Emerg Microbes Infect. 2021;10(1):1530–5.
Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu P, Zhong H, Luo Y, Mei J, et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47(3):845–7.
Guan Y, Wang K, Zeng Y, Ye Y, Chen L, Huang T. Development of a direct and rapid detection method for viable but non-culturable state of Pediococcus acidilactici. Front Microbiol. 2021;12:687691.
Wang J, Gao S, Zhang S, He X, Liu J, Liu A, Li Y, Liu G, Luo J, Guan G, et al. Rapid detection of Babesia motasi responsible for human babesiosis by cross-priming amplification combined with a vertical flow. Parasit Vectors. 2020;13(1):377.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63.
Arfaatabar M, Goodarzi N, Afshar D, Memariani H, Pourmand RI. Rapid detection of mycoplasma pneumoniae by Loop-Mediated Isothermal Amplification (LAMP) in clinical respiratory specimens. Iran J Public Health. 2019;48(5):917–24.
Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin Microbiol Rev. 2017;30(3):747–809.
Xu G, Hu L, Zhong H, Wang H, Yusa S, Weiss TC, Romaniuk PJ, Pickerill S, You Q. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci Rep. 2012;2:246.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This research was supported by the Zhejiang Provincial Medical and Health Technology Project and Medical and Health Technology Project of Hangzhou.
Conflict of interest
Yu Junwei, Zhou Yanqiong, and You Qimin are employed by Ustar Biotechnologies (Hangzhou) Ltd.
Ethics approval and consent to participate
The study of clinical specimens was approved by the Ethics Committee of Hangzhou First People's Hospital (KY-20211203-0127-01). All procedures were conducted according to the Ethical Procedures and Guidelines of the People’s Republic of China.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because they contain information that could compromise research participant privacy/consent.
Code availability
Not applicable.
Consent for publication
Not applicable.
Authors’ contributions
CG: Conceptualization, methodology, investigation, writing original draft, visualization. YJ: Conceptualization, methodology, investigation. CH & CK: Visualization, methodology, data curation. ZY: Methodology, visualization. YQ: Methodology, software. WS: Conceptualization, methodology, data curation, formal analysis, writing original draft. All authors have read and approved the final manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Chen, G., Yu, J., Chen, H. et al. EasyNAT MP Assay: A Simple, Rapid, and Low-Cost Method to Detect Mycoplasma pneumoniae Using Cross-Priming Amplification Technology. Mol Diagn Ther 26, 345–352 (2022). https://doi.org/10.1007/s40291-022-00582-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-022-00582-6