Abstract
Introduction
Breast cancer (BC) is the most frequent malignant disease in women worldwide and is therefore challenging for the healthcare system. Early BC detection remains a leading factor that improves overall outcome and disease management. Aside from established screening procedures, there is a constant demand for additional BC detection methods. Routine BC screening via non-invasive liquid biopsy biomarkers is one auspicious approach to either complete or even replace the current state-of-the-art diagnostics. The study explores the diagnostic potential of urinary exosomal microRNAs with specific BC biomarker characteristics to initiate the potential prospective application of non-invasive BC screening as routine practice.
Methods
Based on a case–control study (69 BC vs. 40 healthy controls), expression level quantification and subsequent biostatistical computation of 13 urine-derived microRNAs were performed to evaluate their diagnostic relevance in BC.
Results
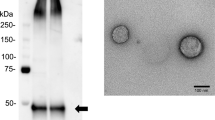
Multilateral statistical assessment determined and repeatedly confirmed a specific panel of four urinary microRNA types (miR-424, miR-423, miR-660, and let7-i) as a highly specific combinatory biomarker tool discriminating BC patients from healthy controls, with 98.6% sensitivity and 100% specificity.
Discussion
Urine-based BC diagnosis may be achieved through the analysis of distinct microRNA panels with proven biomarker abilities. Subject to further validation, the implementation of urinary BC detection in routine screening offers a promising non-invasive alternative in women’s healthcare.




Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. Additional information is available from the corresponding author on reasonable request.
References
GBD 2016 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1211–59. https://doi.org/10.1016/S0140-6736(17)32154-2.
Sharma R. Breast cancer incidence, mortality and mortality-to-incidence ratio (MIR) are associated with human development, 1990–2016: evidence from Global Burden of Disease Study 2016. Breast Cancer. 2019;26(4):428–45.
Momenimovahed Z, Salehiniya H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer (Dove Med Press). 2019;11:151–64.
Oeffinger KC, Fontham ET, Etzioni R, Herzig A, Michaelson JS, Shih YC, et al. Breast Cancer Screening for Women at Average Risk: 2015 guideline update from the American Cancer Society. JAMA. 2015;314(15):1599–614.
Tabar L, Yen AM, Wu WY, Chen SL, Chiu SY, Fann JC, et al. Insights from the breast cancer screening trials: how screening affects the natural history of breast cancer and implications for evaluating service screening programs. Breast J. 2015;21(1):13–20.
Wockel A, Festl J, Stuber T, Brust K, Stangl S, Heuschmann PU, et al. Interdisciplinary screening, diagnosis, therapy and follow-up of breast cancer. Guideline of the DGGG and the DKG (S3-Level, AWMF Registry Number 032/045OL, December 2017)—Part 1 with recommendations for the screening, diagnosis and therapy of breast cancer. Geburtshilfe Frauenheilkd. 2018;78(10):927–48.
Sankaranarayanan R, Swaminathan R, Brenner H, Chen K, Chia KS, Chen JG, et al. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 2010;11(2):165–73.
Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002;225(1):165–75.
Wang L. Early diagnosis of breast cancer. Sensors (Basel). 2017;17(7):E1572. https://doi.org/10.3390/s17071572.
Kudela E, Samec M, Kubatka P, Nachajova M, Laucekova Z, Liskova A, et al. Breast cancer in young women: status quo and advanced disease management by a predictive, preventive, and personalized approach. Cancers (Basel). 2019;11(11):E1791. https://doi.org/10.3390/cancers11111791.
Polivka J Jr, Altun I, Golubnitschaja O. Pregnancy-associated breast cancer: the risky status quo and new concepts of predictive medicine. EPMA J. 2018;9(1):1–13.
Zubor P, Kubatka P, Kajo K, Dankova Z, Polacek H, Bielik T, et al. Why the gold standard approach by mammography demands extension by multiomics? Application of liquid biopsy miRNA profiles to breast cancer disease management. Int J Mol Sci. 2019;20(12):E2878. https://doi.org/10.3390/ijms20122878.
Arneth B. Update on the types and usage of liquid biopsies in the clinical setting: a systematic review. BMC Cancer. 2018;18(1):527.
Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: challenges and opportunities. J Cell Physiol. 2018;233(9):6370–80.
Erbes T, Hirschfeld M, Rucker G, Jaeger M, Boas J, Iborra S, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;28(15):193.
Balacescu O, Petrut B, Tudoran O, Feflea D, Balacescu L, Anghel A, et al. Urinary microRNAs for prostate cancer diagnosis, prognosis, and treatment response: are we there yet? Wiley Interdiscip Rev RNA. 2017;8(6):e1438.
Gasparri ML, Casorelli A, Bardhi E, Besharat AR, Savone D, Ruscito I, et al. Beyond circulating microRNA biomarkers: urinary microRNAs in ovarian and breast cancer. Tumour Biol. 2017;39(5):1010428317695525.
Grayson K, Gregory E, Khan G, Guinn BA. Urine biomarkers for the early detection of ovarian cancer—are we there yet? Biomark Cancer. 2019;11:1179299X19830977.
Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN. Urinary exosomes: the potential for biomarker utility, intercellular signaling and therapeutics in urological malignancy. J Urol. 2016;195(5):1331–9.
Ritter A, Hirschfeld M, Berner K, Rucker G, Jager M, Weiss D, et al. Circulating noncoding RNAbiomarker potential in neoadjuvant chemotherapy of triple negative breast cancer? Int J Oncol. 2020;56(1):47–68.
Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 pathway in cancer. Front Genet. 2017;8:31.
Dufresne S, Rebillard A, Muti P, Friedenreich CM, Brenner DR. A review of physical activity and circulating miRNA expression: implications in cancer risk and progression. Cancer Epidemiol Biomark Prev. 2018;27(1):11–24.
Elghoroury EA, ElDine HG, Kamel SA, Abdelrahman AH, Mohammed A, Kamel MM, et al. Evaluation of miRNA-21 and miRNA Let-7 as prognostic markers in patients with breast cancer. Clin Breast Cancer. 2018;18(4):e721–6.
Masood N, Basharat Z, Khan T, Yasmin A. Entangling relation of micro RNA-let7, miRNA-200 and miRNA-125 with various cancers. Pathol Oncol Res. 2017;23(4):707–15.
Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J Vasc Res. 2009;46(6):527–40.
Thammaiah CK, Jayaram S. Role of let-7 family microRNA in breast cancer. Noncoding RNA Res. 2016;1(1):77–82.
de Anda-Jauregui G, Espinal-Enriquez J, Drago-Garcia D, Hernandez-Lemus E. Nonredundant, highly connected microRNAs control functionality in breast cancer networks. Int J Genomics. 2018;2018:9585383.
Lehmann TP, Korski K, Gryczka R, Ibbs M, Thieleman A, Grodecka-Gazdecka S, et al. Relative levels of let-7a, miR-17, miR-27b, miR-125a, miR-125b and miR-206 as potential molecular markers to evaluate grade, receptor status and molecular type in breast cancer. Mol Med Rep. 2015;12(3):4692–702.
Sun H, Ding C, Zhang H, Gao J. Let7 miRNAs sensitize breast cancer stem cells to radiationinduced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14(4):3285–92.
Bobbili MR, Mader RM, Grillari J, Dellago H. OncomiR-17-5p: alarm signal in cancer? Oncotarget. 2017;8(41):71206–22.
Hesari A, Azizian M, Darabi H, Nesaei A, Hosseini SA, Salarinia R, et al. Expression of circulating miR-17, miR-25, and miR-133 in breast cancer patients. J Cell Biochem. 2019;120:7109–14. https://doi.org/10.1002/jcb.27984.
Li J, Lai Y, Ma J, Liu Y, Bi J, Zhang L, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17(1):745.
Swellam M, Zahran RFK, Abo El-Sadat Taha H, El-Khazragy N, Abdel-Malak C. Role of some circulating MiRNAs on breast cancer diagnosis. Arch Physiol Biochem. 2019;125(5):456–64. https://doi.org/10.1080/13813455.2018.1482355.
Wang Y, Li J, Dai L, Zheng J, Yi Z, Chen L. MiR-17-5p may serve as a novel predictor for breast cancer recurrence. Cancer Biomark. 2018;22(4):721–6.
Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernandez-Losa J, et al. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-alpha, CCNJ, and MEGF9. PLoS One. 2013;8(10):e76247.
Ferracin M, Bassi C, Pedriali M, Pagotto S, D’Abundo L, Zagatti B, et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol Cancer. 2013;12(1):130.
Luo Y, Wang X, Niu W, Wang H, Wen Q, Fan S, et al. Elevated microRNA-125b levels predict a worse prognosis in HER2-positive breast cancer patients. Oncol Lett. 2017;13(2):867–74.
Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chavez J, Trevino V, Rodriguez-Padilla C, et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9(1):53–9.
Amini S, Abak A, Estiar MA, Montazeri V, Abhari A, Sakhinia E. Expression analysis of microRNA-222 in breast cancer. Clin Lab. 2018;64(4):491–6.
Liu S, Wang Z, Liu Z, Shi S, Zhang Z, Zhang J, et al. miR-221/222 activate the Wnt/beta-catenin signaling to promote triple-negative breast cancer. J Mol Cell Biol. 2018;10(4):302–15.
Liang YK, Lin HY, Dou XW, Chen M, Wei XL, Zhang YQ, et al. MiR-221/222 promote epithelial-mesenchymal transition by targeting Notch3 in breast cancer cell lines. NPJ Breast Cancer. 2018;4:20.
Han SH, Kim HJ, Gwak JM, Kim M, Chung YR, Park SY. MicroRNA-222 expression as a predictive marker for tumor progression in hormone receptor-positive breast cancer. J Breast Cancer. 2017;20(1):35–44.
Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang M, et al. Exosome-mediated miR-222 transferring: an insight into NF-kappaB-mediated breast cancer metastasis. Exp Cell Res. 2018;369(1):129–38.
Gao B, Hao S, Tian W, Jiang Y, Zhang S, Guo L, et al. MicroRNA-107 is downregulated and having tumor suppressive effect in breast cancer by negatively regulating brain-derived neurotrophic factor. J Gene Med. 2017;19(12):e2932.
Petrovic N, Davidovic R, Bajic V, Obradovic M, Isenovic RE. MicroRNA in breast cancer: the association with BRCA1/2. Cancer Biomark. 2017;19(2):119–28.
Ai H, Zhou W, Wang Z, Qiong G, Chen Z, Deng S. MicroRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. J Cell Biochem. 2019;120:8696–705. https://doi.org/10.1002/jcb.28157.
Zhang L, Ma P, Sun LM, Han YC, Li BL, Mi XY, et al. MiR-107 down-regulates SIAH1 expression in human breast cancer cells and silencing of miR-107 inhibits tumor growth in a nude mouse model of triple-negative breast cancer. Mol Carcinog. 2016;55(5):768–77.
Li XY, Luo QF, Wei CK, Li DF, Li J, Fang L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7(1):32–40.
Kleivi Sahlberg K, Bottai G, Naume B, Burwinkel B, Calin GA, Borresen-Dale AL, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21(5):1207–14.
Stuckrath I, Rack B, Janni W, Jager B, Pantel K, Schwarzenbach H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6(15):13387–401.
Yang F, Xiao Z, Zhang S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/beta-catenin signaling pathway. Int J Mol Med. 2018;42(6):3355–63.
Huo D, Clayton WM, Yoshimatsu TF, Chen J, Olopade OI. Identification of a circulating microRNA signature to distinguish recurrence in breast cancer patients. Oncotarget. 2016;7(34):55231–48.
Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One. 2012;7(7):e41170.
Zhao H, Gao A, Zhang Z, Tian R, Luo A, Li M, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. 2015;36(6):4763–71.
Smith RA, Jedlinski DJ, Gabrovska PN, Weinstein SR, Haupt L, Griffiths LR. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genom Proteom. 2012;9(3):115–8.
Murria Estal R, Palanca Suela S, de Juan Jimenez I, Egoavil Rojas C, Garcia-Casado Z, Juan Fita MJ, et al. MicroRNA signatures in hereditary breast cancer. Breast Cancer Res Treat. 2013;142(1):19–30.
Wang J, Wang S, Zhou J, Qian Q. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother. 2018;102:147–52.
Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–66.
Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao D, et al. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res Treat. 2015;154(2):423–34.
Shen Y, Ye YF, Ruan LW, Bao L, Wu MW, Zhou Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet Mol Res. 2017;16(1):1–11. https://doi.org/10.4238/gmr16019479.
Krishnan P, Ghosh S, Wang B, Li D, Narasimhan A, Berendt R, et al. Next generation sequencing profiling identifies miR-574-3p and miR-660-5p as potential novel prognostic markers for breast cancer. BMC Genomics. 2015;29(16):735.
Norgen-Biotek-Corp. Total RNA purification kit product insert product #17200, 37500, 17250. 2018. https://norgenbiotek.com/sites/default/files/resources/Total%20RNA%20Purification%20Kit%20Insert%20PI17200-33.pdf
Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014;28(15):29.
Marabita F, de Candia P, Torri A, Tegner J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17(2):204–12.
Tutz G, Binder H. Generalized additive modeling with implicit variable selection by likelihood-based boosting. Biometrics. 2006;62(4):961–71.
R-Core-Team. R: a language and environment for statistical computing. 2018. https://www.r-project.org/
Gerner C, Costigliola V, Golubnitschaja O. Multiomic patterns in body fluids: technological challenge with a great potential to implement the advanced paradigm of 3P medicine. Mass Spectrom Rev. 2019. https://doi.org/10.1002/mas.21612.
Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet. 2013;4:94.
Poel D, Buffart TE, Oosterling-Jansen J, Verheul HM, Voortman J. Evaluation of several methodological challenges in circulating miRNA qPCR studies in patients with head and neck cancer. Exp Mol Med. 2018;50(3):e454.
Djordjevic V, Stankovic M, Nikolic A, Antonijevic N, Rakicevic LJ, Divac A, et al. PCR amplification on whole blood samples treated with different commonly used anticoagulants. Pediatr Hematol Oncol. 2006;23(6):517–21.
Geeurickx E, Hendrix A. Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol Aspects Med. 2019;8:100828.
Duffy D. Standardized immunomonitoring: separating the signals from the noise. Trends Biotechnol. 2018;36(11):1107–15.
Wu L, Qu X. Cancer biomarker detection: recent achievements and challenges. Chem Soc Rev. 2015;44(10):2963–97.
Ghosh RK, Pandey T, Dey P. Liquid biopsy: a new avenue in pathology. Cytopathology. 2019;30(2):138–43.
Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treat. 2017;40(7–8):404–8.
Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–24.
Rossi G, Ignatiadis M. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 2019;79(11):2798–804.
Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95(6):643–60.
Wang HX, Gires O. Tumor-derived extracellular vesicles in breast cancer: from bench to bedside. Cancer Lett. 2019;460:54–64.
Beretov J, Wasinger VC, Millar EK, Schwartz P, Graham PH, Li Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC–MS/MS approach. PLoS One. 2015;10(11):e0141876.
Burton C, Dan Y, Donovan A, Liu K, Shi H, Ma Y, et al. Urinary metallomics as a novel biomarker discovery platform: breast cancer as a case study. Clin Chim Acta. 2016;15(452):142–8.
Guo C, Li X, Ye M, Xu F, Yu J, Xie C, et al. Discriminating patients with early-stage breast cancer from benign lesions by detection of oxidative DNA damage biomarker in urine. Oncotarget. 2017;8(32):53100–9.
Herman-Saffar O, Boger Z, Libson S, Lieberman D, Gonen R, Zeiri Y. Early non-invasive detection of breast cancer using exhaled breath and urine analysis. Comput Biol Med. 2018;1(96):227–32.
Woollam M, Teli M, Angarita-Rivera P, Liu S, Siegel AP, Yokota H, et al. Detection of volatile organic compounds (VOCs) in urine via gas chromatography-mass spectrometry QTOF to differentiate between localized and metastatic models of breast cancer. Sci Rep. 2019;9(1):2526.
Hofbauer SL, de Martino M, Lucca I, Haitel A, Susani M, Shariat SF, et al. A urinary microRNA (miR) signature for diagnosis of bladder cancer. Urol Oncol. 2018;36(12):531 (e1–e8).
Kutwin P, Konecki T, Borkowska EM, Traczyk-Borszynska M, Jablonowski Z. Urine miRNA as a potential biomarker for bladder cancer detection—a meta-analysis. Cent Eur J Urol. 2018;71(2):177–85.
Lekchnov EA, Amelina EV, Bryzgunova OE, Zaporozhchenko IA, Konoshenko MY, Yarmoschuk SV, et al. Searching for the novel specific predictors of prostate cancer in urine: the analysis of 84 miRNA expression. Int J Mol Sci. 2018;19(12):E4088. https://doi.org/10.3390/ijms19124088.
Hung PS, Chen CY, Chen WT, Kuo CY, Fang WL, Huang KH, et al. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12(5):e0177346.
Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86(2):433–44.
Fehlmann T, Ludwig N, Backes C, Meese E, Keller A. Distribution of microRNA biomarker candidates in solid tissues and body fluids. RNA Biol. 2016;13(11):1084–8.
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–41.
Xiao YF, Yong X, Fan YH, Lu MH, Yang SM, Hu CJ. microRNA detection in feces, sputum, pleural effusion and urine: novel tools for cancer screening (Review). Oncol Rep. 2013;30(2):535–44.
Zhou K, Spillman MA, Behbakht K, Komatsu JM, Abrahante JE, Hicks D, et al. A method for extracting and characterizing RNA from urine: for downstream PCR and RNAseq analysis. Anal Biochem. 2017;1(536):8–15.
Koberle V, Pleli T, Schmithals C, Augusto Alonso E, Haupenthal J, Bonig H, et al. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 2013;8(9):e75184.
Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016;49(3):281–303.
Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18(3):371–90.
Wasserstein RL, Schirm AL, Lazar NA. Moving to a world beyond “p < 0.05”. Am Stat. 2019;73(sup1):1–19.
Liu J, Gu Z, Tang Y, Hao J, Zhang C, Yang X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle. 2018;17(3):309–18.
Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-beta signaling pathway. Oncotarget. 2016;7(46):75185–96.
Wu K, Hu G, He X, Zhou P, Li J, He B, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–48.
Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–87.
Xie D, Song H, Wu T, Li D, Hua K, Xu H, et al. MicroRNA424 serves an antioncogenic role by targeting cyclindependent kinase 1 in breast cancer cells. Oncol Rep. 2018;40(6):3416–26.
Shivapurkar N, Vietsch EE, Carney E, Isaacs C, Wellstein A. Circulating microRNAs in patients with hormone receptor-positive, metastatic breast cancer treated with dovitinib. Clin Transl Med. 2017;6(1):37.
Fortunato O, Boeri M, Moro M, Verri C, Mensah M, Conte D, et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis. 2014;11(5):e1564.
He T, Chen P, Jin L, Hu J, Li Y, Zhou L, et al. miR6605p is associated with cell migration, invasion, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2018;17(1):2051–60.
He X, Shu Y. RNA N6-methyladenosine modification participates in miR-660/E2F3 axis-mediated inhibition of cell proliferation in gastric cancer. Pathol Res Pract. 2019;215(6):152393.
Borzi C, Calzolari L, Centonze G, Milione M, Sozzi G, Fortunato O. mir-660-p53-mir-486 network: a new key regulatory pathway in lung tumorigenesis. Int J Mol Sci. 2017;18(1):E222. https://doi.org/10.3390/ijms18010222.
Zhang P, Gao H, Li Q, Chen X, Wu X. Downregulation of microRNA660 inhibits cell proliferation and invasion in osteosarcoma by directly targeting forkhead box O1. Mol Med Rep. 2018;18(2):2433–40.
Zhou C, Chen Z, Dong J, Li J, Shi X, Sun N, et al. Combination of serum miRNAs with Cyfra21-1 for the diagnosis of non-small cell lung cancer. Cancer Lett. 2015;367(2):138–46.
Elkhadragy L, Chen M, Miller K, Yang MH, Long W. A regulatory BMI1/let-7i/ERK3 pathway controls the motility of head and neck cancer cells. Mol Oncol. 2017;11(2):194–207.
Gadducci A, Sergiampietri C, Lanfredini N, Guiggi I. Micro-RNAs and ovarian cancer: the state of art and perspectives of clinical research. Gynecol Endocrinol. 2014;30(4):266–71.
Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107(3):dju492. https://doi.org/10.1093/jnci/dju492.
Qin MM, Chai X, Huang HB, Feng G, Li XN, Zhang J, et al. let-7i inhibits proliferation and migration of bladder cancer cells by targeting HMGA1. BMC Urol. 2019;19(1):53.
Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu YP, et al. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31(4):1715–22.
Yang WH, Lan HY, Tai SK, Yang MH. Repression of bone morphogenetic protein 4 by let-7i attenuates mesenchymal migration of head and neck cancer cells. Biochem Biophys Res Commun. 2013;433(1):24–30.
Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Muller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77(5):1265 (e9–16).
Xiao D, Barry S, Kmetz D, Egger M, Pan J, Rai SN, et al. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016;376(2):318–27.
Chakraborty C, Das S. Profiling cell-free and circulating miRNA: a clinical diagnostic tool for different cancers. Tumour Biol. 2016;37(5):5705–14.
Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan Y, et al. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer. 2017;114:6–11.
Du W, Feng Z, Sun Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochem Biophys Res Commun. 2018;507(1–4):198–202.
Lin H, Lin T, Lin J, Yang M, Shen Z, Liu H, et al. Inhibition of miR-423-5p suppressed prostate cancer through targeting GRIM-19. Gene. 2019;10(688):93–7.
Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinog. 2018;57(9):1223–36.
McDermott AM, Miller N, Wall D, Martyn LM, Ball G, Sweeney KJ, et al. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS ONE. 2014;9(1):e87032.
Ding HX, Lv Z, Yuan Y, Xu Q. MiRNA polymorphisms and cancer prognosis: a systematic review and meta-analysis. Front Oncol. 2018;8:596.
Campos-Parra AD, Mitznahuatl GC, Pedroza-Torres A, Romo RV, Reyes FIP, Lopez-Urrutia E, et al. Micro-RNAs as potential predictors of response to breast cancer systemic therapy: future clinical implications. Int J Mol Sci. 2017;18(6):E1182. https://doi.org/10.3390/ijms18061182.
Liu B, Su F, Chen M, Li Y, Qi X, Xiao J, et al. Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol. 2017;64:44–52.
Xie X, Hu Y, Xu L, Fu Y, Tu J, Zhao H, et al. The role of miR-125b-mitochondria-caspase-3 pathway in doxorubicin resistance and therapy in human breast cancer. Tumour Biol. 2015;36(9):7185–94.
Hong L, Pan F, Jiang H, Zhang L, Liu Y, Cai C, et al. miR-125b inhibited epithelial-mesenchymal transition of triple-negative breast cancer by targeting MAP2K7. Onco Targets Ther. 2016;9:2639–48.
Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–106.
Das R, Gregory PA, Fernandes RC, Denis I, Wang Q, Townley SL, et al. MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res. 2017;77(4):1021–34.
Kong Q, Chen XS, Tian T, Xia XY, Xu P. MicroRNA-194 suppresses prostate cancer migration and invasion by downregulating human nuclear distribution protein. Oncol Rep. 2017;37(2):803–12.
Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao Y, et al. MiRNA-194 activates the Wnt/beta-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;28(385):117–27.
Zhang X, Wei C, Li J, Liu J, Qu J. MicroRNA-194 represses glioma cell epithelialtomesenchymal transition by targeting Bmi1. Oncol Rep. 2017;37(3):1593–600.
Chen Y, Wei H, Liu Y, Zheng S. Promotional effect of microRNA-194 on breast cancer cells via targeting F-box/WD repeat-containing protein 7. Oncol Lett. 2018;15(4):4439–44.
Gilles ME, Slack FJ. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin Ther Targets. 2018;22(11):929–39.
Chernyi VS, Tarasova PV, Kozlov VV, Saik OV, Kushlinskii NE, Gulyaeva LF. Search of MicroRNAs regulating the receptor status of breast cancer in silico and experimental confirmation of their expression in tumors. Bull Exp Biol Med. 2017;163(5):655–9.
Sueta A, Yamamoto Y, Tomiguchi M, Takeshita T, Yamamoto-Ibusuki M, Iwase H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget. 2017;8(41):69934–44.
Zavesky L, Jandakova E, Turyna R, Langmeierova L, Weinberger V, Minar L. Supernatant versus exosomal urinary microRNAs. Two fractions with different outcomes in gynaecological cancers. Neoplasma. 2016;63(1):121–32.
Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8(2):61–81.
Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24(2):48–56.
Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila). 2012;5(3):492–7.
Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom Proteom Bioinform. 2015;13(1):17–24.
Larssen P, Wik L, Czarnewski P, Eldh M, Lof L, Ronquist KG, et al. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol Cell Proteom. 2017;16(3):502–11.
Swellam M, El Magdoub HM, Hassan NM, Hefny MM, Sobeih ME. Potential diagnostic role of circulating MiRNAs in breast cancer: implications on clinicopathological characters. Clin Biochem. 2018;56:47–54.
Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77.
Cui Z, Lin D, Song W, Chen M, Li D. Diagnostic value of circulating microRNAs as biomarkers for breast cancer: a meta-analysis study. Tumour Biol. 2015;36(2):829–39.
Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M, et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8(9):e3045.
Xie S, Wang Y, Liu H, Wang M, Yu H, Qiao Y, et al. Diagnostic significance of circulating multiple miRNAs in breast cancer: a systematic review and meta-analysis. Biomark Med. 2016;10(6):661–74.
Gheinani AH, Vogeli M, Baumgartner U, Vassella E, Draeger A, Burkhard FC, et al. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci Rep. 2018;8(1):3945.
Pimentel F, Bonilla P, Ravishankar YG, Contag A, Gopal N, LaCour S, et al. Technology in microRNA profiling: circulating microRNAs as noninvasive cancer biomarkers in breast cancer. J Lab Autom. 2015;20(5):574–88.
Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8(1):51–60.
Acknowledgements
The authors are particularly grateful for the continuous organizational support and technical assistance given by Mrs. Claudia Nöthling.
Author information
Authors and Affiliations
Contributions
TE, MJ, and MH devised the project outline and experimental study concept. MJ substantially worked out technical details of analytical assays. MJ, DW, and AR performed practical realization of sample handling, processing, and data acquisition. DW and AR were responsible for study-relevant patient data administration. TE, MJ, MH, and DW conducted a collective literature review. GR conceptually designed and overall guided statistical data assessment. GR performed statistical testing to extract decisive informative value. Final data evaluation and determination of conclusive study output were performed by MH, TE, GR, and MJ. MH, TE, and GR wrote the manuscript in consultation with and under critical revision of KB, MJ, DW, and AR. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors TE, MJ, DW, and MH declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest, excepting that they are listed as innovators with respect to a patent submitted to the European Patent Office by the University of Freiburg in 08/2019, with a currently pending patent application status (application No. EP19190989.4). The authors GR, AR, and KB declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
The according investigation protocols (36/12 and 386/16) were approved by the institutional ethical review board of the University of Freiburg. All patients and healthy controls involved provided written informed consent in accordance with the Declaration of Helsinki.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hirschfeld, M., Rücker, G., Weiß, D. et al. Urinary Exosomal MicroRNAs as Potential Non-invasive Biomarkers in Breast Cancer Detection. Mol Diagn Ther 24, 215–232 (2020). https://doi.org/10.1007/s40291-020-00453-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00453-y