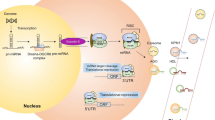
Abstract
Extracellular RNAs consist of coding and non-coding transcripts released from all cell types, which are involved in multiple cellular processes, predominantly through regulation of gene expression. Recent advances have helped us better understand the functions of these molecules, particularly microRNAs (miRNAs). Numerous pre-clinical and human studies have demonstrated that miRNAs are dysregulated in cancer and contribute to tumorigenesis and metastasis. miRNA profiling has extensively been evaluated as a non-invasive method for cancer diagnosis, prognostication, and assessment of response to cancer therapies. Broader applications for miRNAs in these settings are currently under active development. Investigators have also moved miRNAs into the realm of cancer therapy. miRNA antagonists targeting miRNAs that silence tumor suppressor genes have shown promising pre-clinical activity. Alternatively, miRNA mimics that silence oncogenes are also under active investigation. These miRNA-based cancer therapies are in early development, but represent novel strategies for clinical management of human cancer.
Similar content being viewed by others
References
Vlassov AV, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–8.
Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–93.
Gyorgy B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–88.
De Toro J, et al. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203.
An T, et al. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522.
Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9(3–4):358–67.
Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360.
Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62.
Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4.
Lee Y, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70.
Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9(1):112–23.
Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9.
Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60.
Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6.
Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282.
Pigati L, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PloS One. 2010;5(10):e13515.
Ohshima K, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PloS One. 2010;5(10):e13247.
Guduric-Fuchs J, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012;13:357.
Jenjaroenpun P, et al. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201.
Bolukbasi MF, et al. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. Mol Ther Nucl Acids. 2012;1:e10.
Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980.
Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–52.
Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516.
Crescitelli R, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:20677.
Huan J, et al. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73(2):918–29.
Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–8.
Gibbings DJ, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143–9.
Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PloS one. 2008;3(11):e3694.
Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–44.
Feng D, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11(5):675–87.
Fitzner D, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124(Pt 3):447–58.
Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:24641.
Morelli AE, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–66.
Tian T, et al. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111(2):488–96.
Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–4.
Van Deun J, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles. 2014;3:24858.
Yuan T, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413.
Zhou B, et al. Extracellular miRNAs: origin, function and biomarkers in hepatic diseases. J Biomed Nanotechnol. 2014;10(10):2865–90.
Cortez MA, et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77.
Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37(11):460–5.
Zhu C, et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110(9):2291–9.
Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200.
Lai EC. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–4.
Lee I, et al. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009;19(7):1175–83.
Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–5.
Pampalakis G, et al. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem. 2010;43(3):324–7.
Zhu DX, et al. Downregulated Dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci. 2012;103(5):875–81.
Chiosea S, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169(5):1812–20.
Chen WX, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS One. 2014;9(4):e95240.
Rao X, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30(9):1082–97.
Miller TE, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903.
Horie T, et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389(2):315–20.
Koh HJ, et al. Sucrose nonfermenting AMPK-related kinase (SNARK) mediates contraction-stimulated glucose transport in mouse skeletal muscle. Proc Natl Acad Sci USA. 2010;107(35):15541–6.
Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108(22):9232–7.
Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum Mol Genet. 2005;14(22):3371–7.
Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9.
Taverna S, et al. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol Cancer. 2014;13:169.
Kucharzewska P, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci USA. 2013;110(18):7312–7.
Kosaka N, et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288(15):10849–59.
Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. 2014;33:50.
Zhuang G, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–23.
Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer cell. 2014;25(4):501–15.
Rana S, Malinowska K, Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15(3):281–95.
Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–91.
Grange C, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71(15):5346–56.
Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–9.
Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–9.
Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319.
Bahn JH, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–30.
Siomi MC, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–58.
Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113(2):373–80.
Robine N, et al. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol. 2009;19(24):2066–76.
Qiao D, et al. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21(25):3988–99.
Siddiqi S, Terry M, Matushansky I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PloS One. 2012;7(3):e33711.
Lee JH, et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15(2):201–11.
Navarro A, et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget. 2015;6(31):31544–56.
Sun G, et al. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–8.
Grochola LF, et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer. 2008;99(7):1083–8.
Liu C, et al. Combined phenotype of 4 markers improves prognostic value of patients with colon cancer. Am J Med Sci. 2012;343(4):295–302.
He W, et al. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer. 2009;9:426.
Ratajczak J, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20(5):847–56.
Nishikawa R, et al. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21(2):53–6.
Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24.
Dhahbi JM, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013;14:298.
Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138(2):215–9.
Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–68.
Durdevic Z, et al. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013;4(5):931–7.
Gebetsberger J, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909.
Sobala A, Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553–63.
Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–95.
Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1404–9.
Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol. 2015;22(1):11–9.
Marcel V, et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24(3):318–30.
Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89.
Shi X, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66.
Ankerst DP, Thompson IM. Sensitivity and specificity of prostate-specific antigen for prostate cancer detection with high rates of biopsy verification. Arch Ital Urol Androl. 2006;78(4):125–9.
Andriole GL, et al. Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J Natl Cancer Inst. 2012;104(2):125–32.
Schroder FH, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366(11):981–90.
Sharova E, et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br J Cancer. 2016;114(12):1362–6.
Aberle DR, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409.
Halvorsen AR, et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget. 2016.
Hou Y, et al. Appraising MicroRNA-155 as a noninvasive diagnostic biomarker for cancer detection: a meta-analysis. Medicine (Baltimore). 2016;95(2):e2450.
Roth C, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90.
Yamada A, et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015;21(18):4234–42.
Wang S, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136(1):152–61.
Wu J, Li L, Jiang C. Identification and evaluation of serum MicroRNA-29 family for glioma screening. Mol Neurobiol. 2015;52(3):1540–6.
Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Jepsen RK, et al. Intra-tumor heterogeneity of microRNA-92a, microRNA-375 and microRNA-424 in colorectal cancer. Exp Mol Pathol. 2016;100(1):125–31.
Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9(3):189–98.
Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5.
Huang X, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67(1):33–41.
Mangolini A, et al. Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res. 2015;3:12.
Eissa S, et al. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol. 2015;36(6):4487–94.
Toraih EA, et al. Pilot study of serum MicroRNA-21 as a diagnostic and prognostic biomarker in egyptian breast cancer patients. Mol Diagn Ther. 2015;19(3):179–90.
Usmani A, et al. Diagnostic, prognostic and predictive value of MicroRNA-21 in breast cancer patients, their daughters and healthy individuals. Am J Cancer Res. 2015;5(8):2484–90.
Eichelser C, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5(20):9650–63.
Chen W, et al. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour Biol. 2013;34(1):455–62.
Muller V, et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Res Treat. 2014;147(1):61–8.
Meng X, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923–35.
Yoo B, et al. Combining miR-10b-Targeted Nanotherapy with Low-Dose Doxorubicin Elicits Durable Regressions of Metastatic Breast Cancer. Cancer Res. 2015;75(20):4407–15.
Kaboli PJ, et al. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res. 2015;97:104–21.
Pu XX, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–80.
Yamamoto H, et al. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J Gastroenterol. 2012;18(22):2745–55.
Rocci A, et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia. 2014;28(9):1922–6.
Hansen TF, et al. MicroRNA-126 and epidermal growth factor-like domain 7-an angiogenic couple of importance in metastatic colorectal cancer. Results from the Nordic ACT trial. Br J Cancer. 2013;109(5):1243–51.
Garcia V, et al. Extracellular tumor-related mRNA in plasma of lymphoma patients and survival implications. PloS one. 2009;4(12):e8173.
Wu X, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42.
Summerer I, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113(1):76–82.
Huang L, et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011;128(8):1758–69.
Shi XB, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104(50):19983–8.
Nelson PT, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12(2):187–91.
Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610.
Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–89.
Ghosh R, et al. A gold nanoparticle platform for the delivery of functional microRNAs into cancer cells. Biomaterials. 2013;34(3):807–16.
Tivnan A, et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PloS one. 2012;7(5):e38129.
Pecot CV, et al. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67.
Wiggins JF, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30.
Trang P, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–22.
Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–6.
Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15(6):827–35.
Geng Q, et al. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149.
Jiang L, et al. Plasma microRNA might as a potential biomarker for hepatocellular carcinoma and chronic liver disease screening. Tumour Biol. 2015;36(9):7167–74.
Li G, et al. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clin Transl Oncol. 2015;17(9):684–93.
Huang JT, et al. Systematic review and meta-analysis: circulating miRNAs for diagnosis of hepatocellular carcinoma. J Cell Physiol. 2016;231(2):328–35.
Wang RJ, et al. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(1):765–71.
Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–8.
Yue X, et al. Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J Neurosurg. 2016;124(1):122–8.
Chen Y, et al. Plasma miR-15b-5p, miR-338-5p, and miR-764 as Biomarkers for Hepatocellular Carcinoma. Med Sci Monit. 2015;21:1864–71.
Liu N, et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int J Cancer. 2014;134(6):1359–68.
Lin HM, et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014;110(10):2462–71.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors JRT, JZ, DK, and LW disclose no potential conflicts of interest.
Funding
This work was supported by the Advancing a Healthier Wisconsin fund (Project# 5520227) to LW, by the State of Wisconsin Income Tax Check-Off Program in Prostate Cancer Research to DK, by the National Natural Science Foundation of China (grant # 81301752) and China Scholarship Council (CSC) to JZ, and by the Mazie Froedtert Willms & Sue Froedtert Endowed Cancer Fellowship for JRT.
Rights and permissions
About this article
Cite this article
Thompson, J.R., Zhu, J., Kilari, D. et al. Applications of Extracellular RNAs in Oncology. Mol Diagn Ther 21, 1–11 (2017). https://doi.org/10.1007/s40291-016-0239-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-016-0239-7