Abstract
Background
Advanced non-small-cell lung cancer (NSCLC) harbours many genetic aberrations that can be targeted with systemic treatments. Whole-genome sequencing (WGS) can simultaneously detect these (and possibly new) molecular targets. However, the exact added clinical value of WGS is unknown.
Objective
The objective of this study was to determine the early cost effectiveness of using WGS in diagnostic strategies compared with currently used molecular diagnostics for patients with inoperable stage IIIB,C/IV non-squamous NSCLC from a Dutch healthcare perspective.
Methods
A decision tree represented the diagnostic pathway, and a cohort state transition model represented disease progression. Three diagnostic strategies were modelled: standard of care (SoC) alone, WGS as a diagnostic test, and SoC followed by WGS. Treatment effectiveness was based on a systematic review. Probabilistic cost-effectiveness analyses were performed, and threshold analyses (using €80,000 per quality-adjusted life-year [QALY]) was used to explore the early cost effectiveness of WGS.
Results
WGS as a diagnostic test resulted in more QALYs (0.002) and costs (€1534 [incremental net monetary benefit –€1349]), and SoC followed by WGS resulted in fewer QALYs (–0.002) and more costs (€1059 [–€1194]) compared with SoC alone. WGS as a diagnostic test was only cost effective if it was priced at €2000 per patient and identified 2.7% more actionable patients than SoC alone. Treating these additional identified patients with new treatments costing >€4069 per month decreased the probability of cost effectiveness.
Conclusions
Our analysis suggests that providing WGS as a diagnostic test is cost effective compared with SoC followed by WGS and SoC alone if costs for WGS decrease and additional patients with actionable targets are identified. This cost-effectiveness model can be used to incorporate new findings iteratively and to support ongoing decision making regarding the use of WGS in this rapidly evolving field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This analysis suggests that providing whole-genome sequencing (WGS) as a diagnostic test results in more quality-adjusted life-years (QALYs) and costs, that standard of care (SoC) followed by WGS results in fewer QALYs and more costs, and that both strategies are not cost effective compared with SoC alone. |
If costs for sequencing decrease to €2000, WGS as a diagnostic test and SoC followed by WGS are cost effective compared with SoC alone when 2.7 and 4.8% additional patients with actionable targets are detected, respectively. |
1 Introduction
Advanced non-small-cell lung cancer (NSCLC) harbours many genetic aberrations [1,2,3,4] that can be targeted with systemic treatments [4, 5]. Whole-genome sequencing (WGS) is being investigated as a possible molecular diagnostic test to identify druggable targets for NSCLC, given its ability to simultaneously test for all known, and potentially new, molecular targets [6]. However, its added clinical value as a molecular diagnostic test is currently unknown.
Many molecular diagnostic tests exist that can detect a single actionable target, for example, immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) [2, 7]. These tests are often performed sequentially, though this is not ideal given the time required and limited available tissue samples [8, 9]. Next-generation sequencing (NGS), e.g. multi-gene panels, can overcome this problem by simultaneously testing for multiple actionable targets. However, these tests are often restricted to a specific subset of targets and performed in different combinations with other tests according to different hospitals [3, 10, 11]. WGS is a form of NGS that provides information about the whole genome and thus can detect any potential aberration present in the tumour DNA, including those that other techniques focussed only on specific parts of the genome would have missed [6, 12]. However, WGS is expensive, and interpretation of results is complex [11].
The clinical utility of a sequencing technology lies in its ability to accurately identify individuals with a given disease and to aid in personalised and targeted treatment selection. Targeted treatments have demonstrated significant improvement of progression-free survival (PFS) and overall survival (OS) for some patients with NSCLC [13]. Many actionable targets can be tested for (e.g. epidermal growth factor receptor [EGFR], anaplastic lymphoma kinase [ALK], ROS proto-oncogene 1 [ROS1], B-Raf proto-oncogene [BRAF]), and more are still being investigated [4]. Additionally, some aberrations validated in other tumour types (e.g. phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha [PIK3CA], human epidermal growth factor receptor 2 [HER2], and BRCA) also exist in NSCLC but are not yet targetable [3]. With the rapid pace of genomic discoveries and treatment developments, there is a chance that currently used multi-gene panels aimed at selected genes could miss novel targets [14]. Furthermore, adding new targets to panels takes time, and they must be validated. These arguments imply that there is a potential use for WGS as a molecular diagnostic test. However, it is unclear how many actionable targets can be found and how much health gain using WGS would yield over traditional methods. At the same time, the costs for WGS are high but decreasing rapidly, from about €3000 per patient in the Netherlands in 2018 [15] to somewhere between €2500 and €2000 in 2021 (Cuppen E, personal communication). In essence, it is unclear how WGS should be implemented in the diagnostic pathway to provide the most health benefits at an accepted cost [16]. Early cost-effectiveness analysis is a useful tool to address this, as it determines the prerequisites for cost effectiveness and the potential for adoption of a new technology using currently available evidence [17]. To date, only limited evidence of the cost effectiveness of introducing multi-gene panels in molecular diagnostics exists [10, 18, 19]. Only one full economic evaluation of the use of WGS in oncology was identified, and this was specific to incidental findings in colorectal cancer [20]. Economic evaluations of WGS are urgently needed to support its translation and implementation into clinical practice in oncology [19]. Therefore, the objective of this study was to determine the early cost effectiveness of using WGS in diagnostic strategies compared with currently used molecular diagnostics for patients with inoperable stage IIIB,C/IV non-squamous NSCLC, from a Dutch healthcare perspective.
2 Methods
2.1 Overview
A probabilistic decision model was constructed in Microsoft Excel consisting of a decision tree and a state transition model (STM). A societal perspective from the Netherlands was adopted, and the model simulated a cohort with a starting age of 60 years, in 240 one-month cycles over a lifetime horizon.
2.2 Diagnostic Strategies
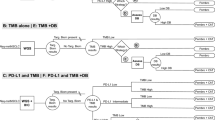
The decision tree is displayed in Fig. 1. Diagnostic strategy A: ‘standard of care’ (SoC) represents the current clinical practice of molecular testing, consisting of an NGS multi-gene panel, FISH, IHC, and Archer fusionPlex, to test for actionable targets. In cases where no actionable target was found, IHC was performed to determine programmed death-ligand 1 (PD-L1) expression levels on the tumour cell’s membrane, which are used in the selection of immunotherapy treatment. Using these tests, the most optimal and least costly testing strategy was created that detected all common actionable targets. This was based on guidelines and an expert’s opinion (PR) [21]. In diagnostic strategy B: ‘WGS as a diagnostic test’, WGS replaced all SoC tests and was used alongside IHC to test for PD-L1 (since WGS cannot detect PD-L1). In diagnostic strategy C: ‘SoC followed by WGS’, patients were first tested with SoC diagnostics and then with WGS if no targets were found. Biopsy and technical success rates were included in all diagnostic strategies, referring to biopsies containing enough tumour cells and the technical performance of the diagnostic test (Fig. 1, Table 1). Where no results were available, patients were considered the ‘target unknown’ subgroup. More details on diagnostics are presented in the Appendix in the electronic supplementary material (ESM).
Diagnostic strategies in inoperable non-small-cell lung cancer. If a biopsy contained too few tumour cells for WGS as a diagnostic test (strategy B), patients could still receive SoC as in strategy A. If a biopsy also contained too few tumour cells for SoC, no test could be performed. If a diagnostic test technically failed twice, no results were available. In case of an unknown target, patients were treated as in the ‘PD-L1 unselected’ subgroup. The circles containing ‘A1’, ‘B1’, and ‘C1’ are truncations of the fully unfolded branch that is only presented the first time it occurs in the decision tree. Target X is a summary of the biomarkers that are currently missed in routine SoC (because of technical and/or analysis challenges) but are reliably detected by WGS. ALK anaplastic lymphoma kinase, BRAF B-Raf proto-oncogene, EGFR epidermal growth factor receptor, IHC immunohistochemistry, KRAS Kirsten rat sarcoma virus proto-oncogene, MET MET proto-oncogene, NSCLC non-small-cell lung cancer, NTRK neurotrophic tropomyosin receptor kinase, PDCT platinum-doublet chemotherapy, PD-L1 programmed death-ligand 1, RET rearranged during transfection proto-oncogene, ROS1 ROS proto-oncogene 1, SoC standard of care, WGS whole-genome sequencing
2.3 Targets and Treatment Selection
The following targets were considered in the model: EGFR mutations (exon 19, 21, T790M, non-classic, and other); ALK rearrangements; ROS1 rearrangements; BRAF (V600) mutations; MET proto-oncogene (MET) (amplifications and exon 14 skipping); rearranged during transfection proto-oncogene (RET) rearrangement; neurotrophic tropomyosin receptor kinase (NTRK) (kinase 1, 2, and 3) gene fusions; KRAS mutation in exon 2, 3, and 4; and PD-L1 expression level. Molecular targets in the aforementioned genes with targeted therapies available in routine practice that are currently missed in SoC (because of technical and/or analysis challenges) but that can be reliably detected with WGS were labelled as ‘target X’ [26]. For the base-case analysis, WGS was assumed to be capable of identifying a proportion of 0.5% target X, based on expert opinion (PR). SoC tests were unable to detect target X because either the test design or the testing strategy was incomplete. All targets in the model were mutually exclusive.
The treatment strategies that were used in the model were a close representation of Dutch clinical practice and were determined through consultation with clinical oncology experts (JM, MP, JA) and the Dutch guidelines [21]. First-line treatments consisted of targeted therapies for the following actionable targets: EGFR, ALK, ROS1, BRAF, and NTRK (including target X). It was assumed that patients with target X were treated with a targeted therapy, summarised as ‘treatment X’. This treatment X was assumed to have the same effectiveness as first-line chemotherapy in patients with EGFR mutations and a cost of €10,000 per patient per model cycle. If no target or a MET, RET, or KRAS aberration was found, first-line treatment was based on PD-L1 and consisted of immunotherapy with or without platinum-doublet chemotherapy (PDCT) in accordance with clinical guidelines [21]. Patients in the unknown target subgroup were treated as PD-L1 unselected. It was assumed that patients switched to second-line treatment after disease progression in the first line. In the second line, patients could receive targeted therapies, immunotherapies with or without PDCT, or chemotherapy, according to the treatment they received in the first line. After disease progression in the second line, patients received best supportive care, which concerned symptom management. Lower detection rates of molecular actionable targets resulted in more patients treated with immunotherapies [21]. Immunotherapy has higher costs and is less effective than targeted therapy. The effectiveness and costs of treatment X will determine the cost effectiveness of WGS, as more patients detected with target X will lead to more patients receiving targeted therapy. Details of the treatment strategies can be found in the Appendix in the ESM.
2.4 Model Structure for Disease Progression
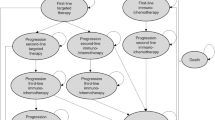
Patients’ disease progression was simulated in the STM with four health states: no progression, progression first line, progression second line, and death (Fig. 2).
State transition model structure. The transition probabilities between the health states are determined by the treatment that was assigned in the decision tree. Note that p4 and p5 were assumed to be equal. p1 probability of progression in the first line (of treatment administration), p2 probability of dying in the first line, p3 probability of progression in the second line, p4 probability of dying in the second line, p5 probability of dying with best supportive care
2.5 Model Input
2.5.1 Identified Targets and Treatment Effects
Parameter values for the decision tree and STM are listed in Table 1. Results from a recent systematic review were used to model the OS and PFS of targeted therapies and immunotherapies in NSCLC [13, 37,38,39,40,41,42,43,44,45,46,47,48,49,50,51]. Treatment effects that were not available in the systematic review were replaced with the OS and PFS of chemotherapy of patients with EGFR mutations. Transition probabilities for first-line treatments were derived from parametric survival models using different distributions chosen according to goodness of fit. Transition probabilities for second-line treatments were derived using exponential distributions, with the assumption of no cycle-dependent transition probabilities in the model. Additionally, the background mortality derived from the Dutch population was applied. More details on model inputs are presented in the Appendix in the ESM.
2.5.2 Costs
Costs and resource use were based on the Dutch costing manual [33], the Dutch healthcare authority [32], and the literature [27]. In the presence of serious adverse events (SAEs) requiring treatment, treatment costs were accounted for. Costs and effects were discounted by a rate of 4.0 and 1.5%, respectively, in line with Dutch guidelines [33]. All costs were based on the average consumer price index of 2020.
2.5.3 Utilities
Health state utilities for the states no progression and progression in the first and second line were obtained from the literature [36]. In the absence of SAEs, health utilities for the three states were assumed to be treatment independent. Disutilities were counted in the presence of treatment-related SAEs. It was assumed that SAEs occurred as soon as the treatment started and disappeared when treated. More details on (dis)utility weights, the studies they were derived from, and SAEs can be found in the Appendix in the ESM.
2.5.4 Model Validation
The model was checked for face validity by consulting clinical oncology and pathology experts (JM, MP, JA, RB, PR, KM), as were all model assumptions. Furthermore, the model was checked for validity using the AdViSHE (Assessment of the Validation Status of Health-Economic decision models) checklist [52] by one of the authors (ZM). The completed checklist is included in the Appendix in the ESM.
2.6 Analyses
2.6.1 Model Output
Model outcomes included costs in €, year 2020 values; life-years (LYs); and quality-adjusted LYs (QALYs) per actionable subgroup. Outcomes were averaged per diagnostic strategy. The incremental cost-effectiveness ratio (ICER) and net monetary benefit were calculated using a willingness-to-pay (WTP) threshold of €80,000 per QALY, as advised for this patient population in the Netherlands [53].
2.6.2 Sensitivity and Threshold Analyses
Deterministic sensitivity analysis (DSA) was performed to test the influence of different parameters on model outcomes. Parameter values were varied based on their standard error or 15% of the mean if not available. Specific parameter values were used for the upper and lower boundaries of the cost for WGS (€2000 [Cuppen E, personal communication] and €3040 [15]) and for the upper boundary of the cost for SoC: €1881 [54]. Probabilistic sensitivity analyses (PSA) were performed to evaluate the parameter uncertainty of all model inputs simultaneously. This was done by drawing random values from pre-specified distributions using Monte Carlo simulations with 2000 iterations. As only the list prices of systemic treatments were available, we performed a scenario analysis in which all treatment costs were halved to reflect the impact of price negotiations between government/hospitals and pharmaceutical companies. One- and three-way threshold analyses were performed to estimate the thresholds for costs for WGS; the proportion of patients with target X identified with WGS; the effectiveness of treatment X (applying the hazard ratio [HR] to the base case); the costs for treatment X; and the costs for SoC for WGS to become cost effective, considering the WTP threshold. A comparison of the strategies ‘WGS as diagnostic test’ and ‘SoC followed by WGS’ versus ‘SoC alone’ was performed in the threshold analyses.
3 Results
3.1 Base-Case Analysis
Table 2 lists the intermediate results of the base-case and scenario sensitivity analysis on the treatment costs and QALYs of the different treatment strategies for the actionable subgroups. Each diagnostic strategy resulted in different proportions of patients within the actionable subgroups. The actionable subgroup resulted in different LYs, QALYs, and costs, which were model averaged per diagnostic strategy. Table 3 lists the resulting overall results of the base-case analysis. SoC followed by WGS was dominated by SoC. WGS as a diagnostic test was not cost effective compared with SoC, considering the WTP threshold. Additional results are listed in Table 8 in the ESM.
3.2 Sensitivity Analyses
Results of the DSA showed that the model results were robust (Fig. 2 in the ESM). Results of the PSA are displayed in Fig. 3 and in Fig. 3 in the ESM. The 95% confidence intervals of both WGS strategies indicated higher costs and uncertainty regarding the QALY gain compared with SoC.
Incremental cost-effectiveness plane of the compared diagnostic strategies. The ellipses represent the 95% confidence interval of 2000 iterations of the diagnostic strategy comparisons. The point estimates represent the mean results of the probabilistic sensitivity analysis. The size of the 95% confidence interval ellipses is explained by the differences between diagnostic strategies caused by parameter uncertainty located in the decision tree of the model. WGS as a diagnostic test had the most differences in diagnostic tests and SoC followed by WGS the least compared with SoC alone. The dotted line represents the willingness-to-pay threshold of €80,000 per QALY. QALY quality-adjusted life-years, SoC standard of care, WGS whole-genome sequencing
WGS as a diagnostic test had a 3.0% probability of being cost effective compared with SoC, and SoC followed by WGS had a 0% probability, given a WTP threshold of €80,000 per QALY.
3.3 Threshold Analyses
One-way threshold analyses showed that the cost for WGS would need to decrease from €2500 to €998 for WGS as a diagnostic test and to €61 for SoC followed by WGS to become cost effective (vs. SoC alone). The proportion of patients with target X would need to increase from 0.5 to 3.8% for WGS as a diagnostic test and to 5.9% for SoC followed by WGS to become cost effective. When assuming that the effect of treatment X on OS and PFS was equal to that of alectinib (applying an HR of 0.21), the WGS strategies were still not cost effective. To become cost effective in each treatment strategy, costs for treatment X would need to decrease to below zero for WGS as a diagnostic test and for SoC followed by WGS. Furthermore, costs for SoC alone would need to increase from €850 to €2334 for WGS as a diagnostic test and to below zero for SoC followed by WGS to become cost effective.
Figure 4 displays the results of the three-way threshold analysis. Figure 4a shows that, if costs for WGS remained at €2500 but treatment X had a better treatment effect on OS and PFS (applying an HR of 0.50), the proportion of patients with target X would need to increase from 0.5 to 7.4% for WGS as a diagnostic test and to 11.4% for SoC followed by WGS to become cost effective. Figure 4b shows that, if costs for treatment X dropped below €4069, whereas the proportion of patients with target X increased from 0.5 to 2.1% for WGS as a diagnostic test and to 3.3% for SoC followed by WGS, a better treatment effect of treatment X resulted in a higher probability of WGS as a diagnostic test and of SoC followed by WGS becoming cost effective. Figure 4c shows that, if costs for WGS remained at €2500, whereas costs for SoC decreased to €619, the proportion of patients with target X would need to increase from 0.5 to 4.3% for WGS as a diagnostic test and to 5.9% for SoC followed by WGS to become cost effective. If costs for SoC increased to €1881, the proportion of patients with target X would need to increase from 0.5 to 1.5% for WGS as a diagnostic test and to 5.9% for SoC followed by WGS to become cost effective. Figure 4d shows that, if costs for treatment X remained at €10,000, whereas costs for WGS decreased to €2000, the proportion of patients with target X would need to increase from 0.5 to 2.7% for WGS as a diagnostic test and to 4.8% for SoC followed by WGS to become cost effective. If costs for WGS increased to €3040, the proportion of patients with target X would need to increase from 0.5 to 5.0% for WGS as a diagnostic test and to 7.1% for SoC followed by WGS to become cost effective.
Three-way threshold analysis of the cost-effectiveness of WGS as a diagnostic test and SoC followed by WGS vs. SoC alone. The lines represent the incremental cost-effectiveness ratio of €80,000 per quality-adjusted life-year for the parameter value combinations of the x- and y-axis and a threshold value for a third parameter that is represented by the different lines. Parameter value combinations that are on the other side of the line in contrast to the area that is indicated with ‘SoC preferred’, result in the cost effectiveness of the other strategies, depending on which line is crossed. The following variables were varied simultaneously: a, cost for WGS, proportion of patients with target X found by WGS and two threshold values for treatment effect of treatment X; b, cost for treatment X, proportion of patients with target X found by WGS and two threshold values for treatment effect of treatment X; c, cost for WGS, proportion of patients with target X found by WGS and two threshold values for cost for SoC; d, cost for treatment X, proportion of patients with target X found by WGS and two threshold values for cost for WGS. HR hazard ratio, SoC standard of care, WGS whole-genome sequencing
4 Discussion
The early cost effectiveness of using WGS in diagnostic strategies compared with SoC to identify targeted therapies for patients with inoperable NSCLC was determined. This model used a Dutch healthcare perspective and was informed by inputs derived from the literature. Our findings show that both diagnostic strategies including WGS were not cost effective compared with most optimal SoC. The most efficient use of WGS appears to be as a diagnostic test for all patients, supported by SoC diagnostics in cases where the biopsy contains too few tumour cells. Using WGS as a diagnostic test resulted in a modest increase in QALYs (0.002) and higher costs (€1534) per patient. This was mainly because of the few additional targets in the model that could be treated with cost-effective targeted therapies. For example, we found that WGS as a diagnostic test could become cost effective if costs decreased to €2000 and 2.7% additional patients with actionable targets for routine targeted therapy were identified (assuming treatment X costs €10,000 per month). However, this is dependent on the cost effectiveness of treatment(s) targeted at the additional targets identified with WGS. If costs for WGS remained at €2500, identifying 2.7% more patients eligible for targeted therapy could result in cost effectiveness if monthly treatment costs to treat these additional patients decreased substantially (31.1%).
It has been suggested that there are more targets available or under investigation in incurable cancers that are not detected by SoC and that can be targeted with systemic treatments [3, 6, 11, 26, 55, 56]. Additionally, WGS can detect non-hotspot targets within known genes, e.g. EGFR, that are missed by SoC and can be treated. However, these data are immature and not yet validated so were not incorporated in current analyses. If the number of actionable targets eventually increases, simultaneous testing using NGS techniques will likely be the most optimal strategy in terms of costs [3, 10, 18], probably because more (cost-)effective targeted therapies become available over time [10]. In many cases, newly found targets have led to off-label treatments or use as experimental drugs in clinical trial settings. This is expensive and not always accessible to all potentially eligible patients [6, 55, 56]. Therefore, finding more actionable targets with WGS does not necessarily lead to a more efficient use of healthcare resources [10], although benefits for patients might increase [6]. Additionally, the ‘added value’ of WGS goes beyond the direct health benefits we captured in our analysis. For example, the scope of using WGS data for treatment decision support could be extended to an earlier disease stage, allowing for previous WGS results to be used for selecting novel treatments that emerge for future patients [57]. The value of WGS could encapsulate wider public benefits, workability, macro-economic value for diagnostic laboratories, and other social factors [57]. However, these values cannot always be easily assessed in health economic evaluations.
A strength of this research is that the diagnostic pathways were defined based on guidelines and extensive clinical expert consultation. We analysed two different positions of WGS in the diagnostic pathway and performed threshold analyses on both strategies. This enabled us to explore many different scenarios and analyse their impact on the early cost effectiveness of WGS. This can be further extended to new scenarios that can help us better understand what to focus future research on to reduce uncertainty. We created a detailed model that can be easily adjusted when new data become available. In addition, we believe that, with adjustments, the model is also capable of simulating other forms of advanced cancers beyond NSCLC.
A limitation of this research is the inability of a cohort STM to properly capture the complexity of personalised medicine. In this context, it is often better to use a patient-level model. However, such a model may have only limited advantages regarding implementing multiple time scales in an efficient way. In this early stage, understanding how to use WGS as diagnostic test should be investigated first. This entails capturing the boundaries of essential parameter values, which can also be done with a cohort model. Because of the limited data, we had to make some model assumptions. The most influential assumption was regarding the treatment effectiveness of some therapies, most notably of treatment X. We also assumed a most optimal SoC strategy in which all the standard routine biomarkers are being tested with a comprehensive multi-gene panel at the DNA and RNA level. If the SoC strategy was not as optimal (which is most likely when using real-world data), diagnostic costs would likely increase, and actionable targets could be missed. However, we did explore the impact of most assumptions with extensive sensitivity analyses and by consulting clinical experts. Costs for keeping SoC tests up to date and delays caused by required technical adaptations and validations were not included in our model.
To our knowledge, this research provides the first set of evidence of the most optimal use of WGS as a diagnostic test from a health economic perspective. Our findings provide direction for future research into and design of the diagnostic pathway for WGS. Optimising (adapting and updating) SoC diagnostics to reduce costs and increase efficiency could be considered but requires time and resources. In contrast, optimising WGS is less often necessary and requires fewer resources, as it provides information about the whole genome and concerns only one test. Additionally, we provided threshold values of several factors that need to be achieved to make future research worthwhile. Future research should focus on the clinical value of WGS, specifically understanding how to use genomic information for treatment decision support and biomarker development. Furthermore, novel predictive biomarkers for clinical response to immunotherapy are needed and could lead to more health gains and cost savings [5]. Currently, only a subset of patients experiences clinical benefit from immunotherapy, resulting in over treatment with expensive therapies.
5 Conclusions
Our analysis suggests that providing WGS as a diagnostic test would be cost effective compared with SoC followed by WGS and SoC alone if costs for WGS decreased and additional patients with actionable targets were identified. This cost-effectiveness model can be used to incorporate new findings iteratively and to support ongoing decision making regarding the use of WGS in this rapidly evolving field.
References
Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19(9):495–509.
Morgensztern D, Campo MJ, Dahlberg SE, Doebele RC, Garon E, Gerber DE, et al. Molecularly targeted therapies in non-small-cell lung cancer annual update 2014. J Thoracic Oncol. 2015;10(1 Suppl 1):S1–63.
Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–505.
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311.
Assi HI, Kamphorst AO, Moukalled NM, Ramalingam SS. Immune checkpoint inhibitors in advanced non-small cell lung cancer. Cancer. 2018;124(2):248–61.
van der Velden DL, Hoes LR, van der Wijngaart H, van Berge Henegouwen JM, van Werkhoven E, Roepman P, et al. The drug rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature. 2019;574(7776):127–31.
Sholl LM, Aisner DL, Varella-Garcia M, Berry LD, Dias-Santagata D, Wistuba II, et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the lung cancer mutation consortium experience. J Thorac Oncol. 2015;10(5):768–77.
Dong L, Wang W, Li A, Kansal R, Chen Y, Chen H, et al. Clinical next generation sequencing for precision medicine in cancer. Curr Genomics. 2015;16(4):253–63.
Popper HH, Tímár J, Ryska A, Olszewski W. Minimal requirements for the molecular testing of lung cancer. Transl Lung Cancer Res. 2014;3(5):301–4.
Steuten L, Goulart B, Meropol NJ, Pritchard D, Ramsey SD. Cost effectiveness of multigene panel sequencing for patients with advanced non-small-cell lung cancer. JCO Clin Cancer Inform. 2019;3:1–10.
Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121(4):631–9.
Katsila T, Patrinos GP. Whole genome sequencing in pharmacogenomics. Front Pharmacol. 2015;6:61.
Simons M, Ramaekers B, Peeters A, Mankor J, Paats M, Aerts J, et al. Observed versus modelled lifetime overall survival of targeted therapies and immunotherapies for advanced non-small cell lung cancer patients - a systematic review. Crit Rev Oncol Hematol. 2020;153:103035.
Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1(4):466–74.
Pasmans CTB, Tops BBJ, Steeghs EMP, Coupé VMH, Grünberg K, de Jong EK, et al. Micro-costing diagnostics in oncology: from single-gene testing to whole- genome sequencing. Expert Rev Pharmacoecon Outcomes Res. 2021;6:1–11.
Schwarze K, Buchanan J, Fermont JM, Dreau H, Tilley MW, Taylor JM, et al. The complete costs of genome sequencing: a microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet Med. 2020;22(1):85–94.
Briggs AH, Weinstein MC, Fenwick EA, Karnon J, Sculpher MJ, Paltiel AD. Model parameter estimation and uncertainty analysis: a report of the ISPOR-SMDM Modeling Good Research Practices Task Force Working Group-6. Med Decis Making. 2012;32(5):722–32.
Tan AC, Lai GGY, Tan GS, Poon SY, Doble B, Lim TH, et al. Utility of incorporating next-generation sequencing (NGS) in an Asian non-small cell lung cancer (NSCLC) population: Incremental yield of actionable alterations and cost-effectiveness analysis. Lung Cancer. 2020;139:207–15.
Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–30.
Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17(7):587–95.
FederatieMedischSpecialisten. Richtlijnendatabase. 2020. https://richtlijnendatabase.nl/richtlijn/niet_kleincellig_longcarcinoom/systemische_behandeling_stadium_iv_nsclc/behandeling_pati_nten_met_een_zeldzame_mutatie_bij_nsclc.html. Accessed 17 Jun 2021.
IKNL. NKR cijfers/IKNL. 2019. https://www.iknl.nl/. Accessed 2 Jul 2020.
Kuijpers C, Hendriks LEL, Derks JL, Dingemans AC, van Lindert ASR, van den Heuvel MM, et al. Association of molecular status and metastatic organs at diagnosis in patients with stage IV non-squamous non-small cell lung cancer. Lung Cancer. 2018;121:76–81.
Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7(6):596–609.
Dietel M, Savelov N, Salanova R, Micke P, Bigras G, Hida T, et al. Real-world prevalence of programmed death ligand 1 expression in locally advanced or metastatic non-small-cell lung cancer: the global, multicenter EXPRESS study. Lung Cancer. 2019;134:174–9.
Monkhorst K, Samsom K, Schipper L, Roepman P, Bosch L, de Bruijn E, et al. Validation of whole genome sequencing in routine clinical practice. Ann Oncol. 2020;2020(19):31.
(ZIN) DNHCI. Drug costs [in Dutch: Medicijnkosten]. 2016. https://www.medicijnkosten.nl/. Accessed 12 Mar 2020.
(ZIN) DNHCI. Farmacotherapeutisch kompas. 2016. https://www.farmacotherapeutischkompas.nl/. Accessed 12 Mar 2020.
Holleman MS, Al MJ, Zaim R, Groen HJM, Uyl-de Groot CA. Cost-effectiveness analysis of the first-line EGFR-TKIs in patients with non-small cell lung cancer harbouring EGFR mutations. Eur J Health Econ. 2020;21(1):153–64.
Aguiar PN Jr, Haaland B, Park W, San Tan P, Del Giglio A, de Jr. Lima LG. Cost-effectiveness of osimertinib in the first-line treatment of patients with EGFR-mutated advanced non-small cell lung cancer. JAMA Oncol. 2018;4(8):1080–4.
van der Linden N, Bongers ML, Coupé VM, Smit EF, Groen HJ, Welling A, et al. Costs of non-small cell lung cancer in the Netherlands. Lung Cancer. 2016;91:79–88.
(NZa) DHA. DBC tariff application [in Dutch: DBC zorgproducten tariefapplicatie]. 2016. Accessed 14 Mar 2020.
Hakkaart-van Roijen LVN, Boumans CAM, Kanters T, Tan SS. Costing manual [in Dutch: Kostenhandleiding]. 2016.
Louie AV, van Werkhoven E, Chen H, Smit EF, Paul MA, Widder J, et al. Patient reported outcomes following stereotactic ablative radiotherapy or surgery for stage IA non-small-cell lung cancer: Results from the ROSEL multicenter randomized trial. Radiother Oncol. 2015t;117(1):44–8.
van Baal PHM, Wong A, Slobbe LCJ, Polder JJ, Brouwer WBF, de Wit GA. Standardizing the inclusion of indirect medical costs in economic evaluations. PharmacoEconomics. 2011;29(3):175–87.
Chouaid C, Agulnik J, Goker E, Herder GJ, Lester JF, Vansteenkiste J, et al. Health-related quality of life and utility in patients with advanced non-small-cell lung cancer: a prospective cross-sectional patient survey in a real-world setting. J Thorac Oncol. 2013;8(8):997–1003.
Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–34.
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15(2):213–22.
Gridelli C, Ciardiello F, Gallo C, Feld R, Butts C, Gebbia V, et al. First-line erlotinib followed by second-line cisplatin-gemcitabine chemotherapy in advanced non-small-cell lung cancer: the TORCH randomized trial. J Clin Oncol. 2012;30(24):3002–11.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46.
Wu YL, Zhou C, Liam CK, Wu G, Liu X, Zhong Z, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label ENSURE study. Ann Oncol. 2015;26(9):1883–9.
Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol. 2015;26(9):1877–83.
Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol. 2011;29(21):2866–74.
Mok TSK, Kim SW, Wu YL, Nakagawa K, Yang JJ, Ahn MJ, et al. Gefitinib plus chemotherapy versus chemotherapy in epidermal growth factor receptor mutation-positive non-small-cell lung cancer resistant to first-line Gefitinib (IMPRESS): overall survival and biomarker analyses. J Clin Oncol. 2017;35(36):4027–34.
Soria JC, Wu YL, Nakagawa K, Kim SW, Yang JJ, Ahn MJ, et al. Gefitinib plus chemotherapy versus placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): a phase 3 randomised trial. Lancet Oncol. 2015;16(8):990–8.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–25.
Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377(9):829–38.
Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–92.
Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–50.
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–30.
Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–33.
Vemer P, Corro Ramos I, van Voorn GA, Al MJ, Feenstra TL. AdViSHE: a validation-assessment tool of health-economic models for decision makers and model users. Pharmacoeconomics. 2016;34(4):349–61.
Versteegh MM, Ramos IC, Buyukkaramikli NC, Ansaripour A, Reckers-Droog VT, Brouwer WBF. Severity-adjusted probability of being cost effective. PharmacoEconomics. 2019;37(9):1155–63.
van de Ven M, Koffijberg H, Retèl V, Monkhorst K, Smit E, van Harten W, et al. Real-world utilization of biomarker testing for patients with advanced non-small-cell lung cancer in a tertiary referral center and referring hospitals. J Mol Diagn. 2021;23(4):484–94.
Roepman P, Bosch L, Samsom K, Schipper L, de Bruijn E, Hoes L, et al. 1408P—WGS implementation in standard cancer diagnostics for every cancer patient (WIDE). Ann Oncol. 2019;30:v575.
Trédan O, Wang Q, Pissaloux D, Cassier P, de la Fouchardière A, Fayette J, et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30(5):757–65.
Simons M, Van De Ven M, Coupé V, Joore M, IJzerman M, Koffijberg E, Frederix G, Uyl-De Groot C, Cuppen E, Van Harten W, Retèl V. Early technology assessment of using whole genome sequencing in personalized oncology. Expert Rev Pharmacoecon Outcomes Res. 2021;21(3):343–51. https://doi.org/10.1080/14737167.2021.1917386.
Simons M, Ramaekers B, Retèl V, Joore M. Early cost-effectiveness model of whole genome sequencing as a molecular diagnostic test versus standard of care in locally advanced and metastatic (Stage IIIB,C/ IV) non-squamous non-small cell lung cancer. 2021. https://doi.org/10.5281/zenodo.5112062.
Acknowledgements
The authors thank Dr. Kim Monkhorst from the Netherlands Cancer Institute-Antoni van Leeuwenhoek hospital for providing expert opinion within the field of clinical pathology. The authors also thank Ms. Danalyn Byng from the Netherlands Cancer Institute-Antoni van Leeuwenhoek hospital, for providing English language editing and review support. Furthermore, the authors thank all members of the Technology Assessment of Next Generation Sequencing in Personalized Oncology (TANGO) consortium (https://zenodo.org/communities/tango-wgs/?page=1&size=20) and the funding organisations the Netherlands Organisation for Health Research and Development (ZonMw) (grant numbers 846001002), the Dutch Cancer Society (KWF), and the Dutch healthcare insurance company Zilveren kruis Achmea for making this research possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the Netherlands Organisation for Health Research and Development (ZonMw) (grant number 846001002), the Dutch Cancer Society (KWF), and the Dutch healthcare insurance company Zilveren kruis Achmea. The funding sources had no involvement in the conduct of this research. The open access fee was paid by the Maastricht University Medical Centre and the Maastricht University.
Conflicts of Interest
Bram Ramaekers, Rogier Butter, Joanne Mankor, Marthe Paats, Paul Roepman, Carin Uyl-de Groot, Wim van Harten, and Manuela Joore have no conflicts of interest that are directly relevant to the content of this article. Martijn Simons, Zakile Mfumbilwa, and Veerle Coupé have received grants from governmental funding of medical research by ZonMw. Valesca Retèl has received grants from Agendia B.V. and Intuitive Surgical outside the submitted work. Joachim Aerts has received personal fees and non-financial support from Biocad outside the submitted work; holds stock options with Amphera; and has a patent in allogenic tumour cell lysate licensed to Amphera, combination immunotherapy in cancer pending, and biomarker for immunotherapy pending.
Availability of Data and Materials
All data generated or analysed during this study are included in this published article and its supplementary information files. The model created during the current study is available in the Zenodo repository [https://doi.org/10.5281/zenodo.5112062] [58].
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Code availability
The used model was created in Microsoft Excel.
Authors’ Contributions
MS: Conceptualisation and design, data acquisition, methodology, formal analysis and interpretation of the data, investigation, validation, writing original draft, writing review and editing, and critical revision of the article for important intellectual content. VR and MJ: Conceptualisation and design, methodology, formal analysis and interpretation of the data, writing original draft, supervision, writing review and editing, and critical revision of the article for important intellectual content. BR: Conceptualisation and design, methodology, formal analysis and interpretation of the data, writing original draft, writing review and editing, and critical revision of the article for important intellectual content. RB, JM, MP, JA, PR: Validation and interpretation of the data, writing review and editing, and critical revision of the article for important intellectual content. ZM and VC: Methodology, validation and interpretation of the data, writing review and editing, and critical revision of the article for important intellectual content. CUG: Interpretation of the data, writing review and editing, and critical revision of the article for important intellectual content. WH: Interpretation of the data, writing review and editing, supervision, and critical revision of the article for important intellectual content.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Simons, M.J.H.G., Retèl, V.P., Ramaekers, B.L.T. et al. Early Cost Effectiveness of Whole-Genome Sequencing as a Clinical Diagnostic Test for Patients with Inoperable Stage IIIB,C/IV Non-squamous Non-small-Cell Lung Cancer. PharmacoEconomics 39, 1429–1442 (2021). https://doi.org/10.1007/s40273-021-01073-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40273-021-01073-y