Abstract
Background
Elimination of rosiglitazone in humans is via hepatic metabolism. The existing studies suggest that CYP2C8 is the major enzyme responsible, with a minor contribution from CYP2C9; however, other studies suggest the involvement of additional cytochrome P450 enzymes and metabolic pathways. Thus a full picture of rosiglitazone metabolism is unclear.
Objective
This study aimed to improve the current understanding of potential drug–drug interactions and implications for therapy by evaluating the kinetics of rosiglitazone metabolism and examining the impact of specific inhibitors on its metabolism using the substrate depletion method.
Methods
In vitro oxidative metabolism of rosiglitazone in human liver microsomes obtained from five donors was determined over a 0.5–500 µM substrate range including the contribution of CYP2C8, CYP2C9, CYP3A4, CYP2E1, and CYP2D6.
Results
The maximum reaction velocity was 1.64 ± 0.98 nmol·mg−1·min−1. The CYP2C8 (69 ± 20%), CYP2C9 (42 ± 10%), CYP3A4 (52 ± 23%), and CEP2E1 (41 ± 13%) inhibitors all significantly inhibited rosiglitazone metabolism.
Conclusion
The results suggest that other cytochrome P450 enzymes, including CYP2C9, CYP3A4, and CEP2E1, in addition to CYP28, also play an important role in the metabolism of rosiglitazone. This example demonstrates that understanding the complete metabolism of a drug is important when evaluating the potential for drug–drug interactions and will assist to improve the current therapeutic strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
In this study, a more comprehensive picture of rosiglitazone metabolism was demonstrated. |
The work presented here will assist in the better management of rosiglitazone use in regard to polypharmacy and pharmacogenetics. |
1 Introduction
Rosiglitazone belongs to the synthetic thiazolidinedione class of drugs that improve insulin sensitivity in humans, and has been used as a potent glucose-lowering medicine in the treatment of type 2 diabetes [1–3]. Elimination of rosiglitazone in humans is mainly metabolic [4] via N-demethylation and para-hydroxylation of the pyridine ring [4] and has been reported to be the same in rats and dogs as in humans [5, 6]. All studies to date suggest that phase II metabolism conjugates the phase I metabolites by sulphation and glucuronidation, and that there is no direct conjugation of rosiglitazone [4].
It has previously been reported that CYP2C8 is the major enzyme responsible for rosiglitazone metabolism in humans, with a minor contribution from CYP2C9 [6]. However, all existing studies of rosiglitazone metabolism have been undertaken using the metabolite formation method, in which only the formation of N-desmethyl rosiglitazone and para-hydroxy rosiglitazone were considered [6]. While formation of these two metabolites has been consistently reported in all 47 human liver microsome samples screened by the Baldwin group [6], in vivo studies have demonstrated that N-desmethyl- and para-hydroxy-related metabolites of rosiglitazone (including their conjugate forms) account for only ~60% of rosiglitazone eliminated in urine and feces [4]. This suggests that ~40% of rosiglitazone is eliminated in the form of metabolites that are not related to N-demethylation and para-hydroxylation pathways, and that other metabolic pathways and/or other cytochrome P450 enzymes also make a major contribution to rosiglitazone metabolism [4]. Furthermore, in an in vivo study, ketoconazole, a potent inhibitor of CYP3A4 [7], increased the plasma concentration of rosiglitazone by ~47% [8]. Therefore, it appears that a complete picture of rosiglitazone metabolism is yet to be established.
The substrate depletion approach to investigating drug metabolism, unlike the metabolite formation method used in existing studies, provides the opportunity to investigate the total cytochrome P450-mediated metabolism of rosiglitazone without complete knowledge of the metabolites or access to the known metabolites. This approach is particularly valuable for obtaining a more complete picture of the kinetics of a substrate’s metabolism, since the sum of the activity of all of the enzymes and all the metabolic pathways that contribute to the metabolism of the parent drug are considered in the evaluation of enzyme kinetics. Therefore, the aim of this study was to examine the in vitro metabolism of rosiglitazone and examine the impact of specific inhibitors on its metabolism in human liver microsomes using the substrate depletion method.
2 Materials and Methods
2.1 Chemicals and Reagents
Rosiglitazone maleate was purchased from Selleck Chemicals LLC (TX, USA); acetonitrile was purchased from Scharlau Chemie S.A. La. Jota (Barcelona, Spain). Magnesium sulfate was purchased from Sigma (Steinheim, Germany). Montelukast was purchased from Sapphire Bioscience (Beaconsfield, NSW, Australia). DL-Isocitric acid, isocitric dehydrogenase, β-nicotinamide adenine dinucleotide phosphate sodium salt, furafylline, quinidine, 13-cis retinoic acid, diethyldithiocarbamate, and sulfaphenazole were purchased from Sigma-Aldrich (Castel Hill, NSW, Australia). Ketoconazole and troleandomycin were purchased from BIOTREND Chemikalien GmbH (Koln, Germany). Human liver microsomes were purchased from Celsis (In Vitro Technologies, Brussels, Belgium). Detailed information on these human liver microsomes is provided in Table 1.
Milli-Q water was prepared using a Milli-Q Ultrapure Water System (Millipore, USA).
2.2 Microsomal Incubation
In this study, a substrate depletion method was used. The protein concentrations and incubation times for rosiglitazone metabolism in the microsomes were optimized to ensure that the metabolism of rosiglitazone at the incubation times and protein concentrations used in the final experiment followed first-order kinetics. Rosiglitazone metabolism in human liver microsomes (Table 1) was performed in the incubation media using sodium phosphate buffer (100 mM, pH 7.4), with a final volume of 100 μL containing: 40 μg microsomal protein in 50 μL of buffer (equal to 0.4 mg/mL), 25 μL of the NADPH-generating system (including 7 mM NADP, 1.25 units isocitric acid dehydrogenase, 18 mM isocitric acid, and 20 mM magnesium chloride) [9], and 25 μL rosiglitazone (at required concentrations). Incubations were performed at 37 °C in a shaking water bath for 0–25 min, with sampling times of 0, 5, 7.5, 10, 15, 20, and 25 min. The concentration of the remaining rosiglitazone in the supernatants was measured using the analytical method described in Sect. 2.4.
The kinetics of rosiglitazone metabolism were studied at the following rosiglitazone concentrations: 0.5, 2.5, 5, 10, 20, 40, 60, 80, 100, 200, 350, and 500 µM.
2.3 Inhibition Studies with Chemical Inhibitors
The assessment of the impact of specific P450 enzymes believed to be involved in the bio-transformation of rosiglitazone in humans was performed using two different sets of experiments.
One set of inhibition studies aimed to examine the impact of inhibition of P450 enzymes believed to be involved in the bio-transformation of rosiglitazone (CYP2C8 and CYP2C9) on metabolism kinetics. These experiments were performed across the full substrate concentration range between 0.5 and 500 µM using the same incubation conditions as described above. In this set of experiments, 40 μg microsomal protein in 25 μL of buffer (instead of in 50 μL of buffer) was used, thus allowing the addition of the inhibitor (25 μL), and the final volume of the incubation media remained the same at 100 μL. These inhibition studies were performed using 0.02 µM montelukast (CYP2C8) [10] and 2.5 µM sulfaphenazole (CYP2C9) [11].
The second set of inhibition studies aimed to screen the impact of specific P450 enzyme inhibitors other than CYP2C8 and CYP2C9 in the metabolism of rosiglitazone. These experiments were performed at a rosiglitazone concentration of 10 µM in triplicate, with the same incubation conditions as above. These inhibition studies were performed using 0.02 µM montelukast (CYP2C8), 2.5 and 10 µM sulfaphenazole (CYP2C9), 1 µM ketoconazole (CYP3A4) [6], 10 µM furafylline (CYP1A2) [6], 1 µM quinidine (CYP2D6) [6], 142 µM 13-cis retinoic acid (CYP2C8) [6], 40 µM troleandomycin (CYP3A4) [6], and 40 µM diethyldithiocarbamate (CYP2E1) [12], as well as both montelukast 0.02 µM (CYP2C8) and sulfaphenazole 2.5 µM (CYP2C9) in combination.
All inhibitions were performed at 37 °C in a shaking water bath for 0–25 min, with sampling times of 0, 5, 7.5, 10, 15, 20, and 25 min. In the inhibition study with furafylline, troleandomycin, and diethyldithiocarbamate, there was a further 10-min pre-incubation before adding rosiglitazone and initiation of sampling at the abovementioned time points [6, 12]. Appropriate controls for the inhibition studies with the final methanol concentrations above 0.1% [13] were performed in individual human liver microsomes.
2.4 Quantification of Rosiglitazone in Incubated Microsomal Samples
The concentrations of rosiglitazone in incubated samples were determined using the high-performance liquid chromatography (HPLC) assay described in detail previously [14]. Briefly, the microsomal reaction was stopped by the addition of 300 μL of ice cold acetonitrile. The samples were then vortexed briefly before centrifugation at 16,200 rpm (12,200×g) for 20 min at 4 °C to separate the precipitated protein and supernatant. Then 20 μL of this supernatant was injected onto the HPLC system, and the concentration of the remaining rosiglitazone in the supernatants was measured using the HPLC assay. The detection of peaks was performed by using the fluorescence detector set at excitation and emission wavelengths 247 and 367 nm, respectively. The range of standard curve calibration was between 0.15 and 10 µM. Dilutions of one in 20 or one in 50 were applied after precipitation for samples with higher concentrations of rosiglitazone than those within the calibration curve range. The incubated samples with substrate concentrations of 20, 40, 60, 80, and 100 µM were diluted one in 20 with the mobile phase before injection into the HPLC. Samples with substrate concentrations of 200, 350, and 500 µM were diluted one in 50 with mobile phase before injection into the HPLC system.
2.5 Data Analysis and Determination of Enzyme Kinetics
The depletion rate constant (K dep) of rosiglitazone metabolism at each substrate concentration was calculated from the Ln concentration versus time profile, and then the concentration of rosiglitazone at time zero (C 0) was calculated. These were determined from the velocity of rosiglitazone metabolism in the microsomes over the full substrate range of 0.5–500 µM. To do this, the concentration of remaining rosiglitazone at each time point was transformed logarithmically (Ln concentration) and the slope (K dep) and the intercept of the Ln concentration over time were estimated via linear regression of Ln concentration over time. C 0 was then calculated from the intercept as the exponent of the intercept. Then the initial reaction velocity at time zero (v) at each substrate concentration was calculated from K dep and C 0 using Equation (1). The reaction velocity at each substrate concentration was expressed as nmol·mg−1·min−1. Microsoft Excel (2007) was used for the calculations of K dep, C 0, and v.
Equation 1: Calculating reaction velocity using K dep and C 0:
To evaluate the enzyme kinetics of rosiglitazone metabolism, single enzyme kinetic models (Equations 2, 3, and 4) were fitted to each reaction velocity profile (velocity/substrate concentration).
Equation 2: Michaelis–Menten kinetics (MM, with two parameters):
Equation 3: Michaelis–Menten with substrate inhibition kinetics (MM,IS, with three parameters):
Equation 4: Allosteric (cooperative) kinetics with three parameters:
where V max is the maximum velocity; K m is the substrate concentration at which the reaction velocity is 50% of V max; S is substrate concentration; K i is the constant indicating the degree of substrate inhibition; n is equivalent to the allosteric coefficient for cooperative substrate binding; K prime (K′) is the substrate concentration at which the reaction velocity equals 50% of V max (equivalent to K m derived by the MM equation).
All fitting of the models was performed using Prism software (GraphPad Prism v5.01, GraphPad Software, Inc. CA, USA). The most appropriate enzyme kinetic model was selected according to the F test. Where the F test could not calculate the best fit because of equal parameters between two tested models, Akaike’s information criterion (AIC) was used to determine the most appropriate enzyme kinetic model.
The inhibition data at a rosiglitazone concentration of 10 µM were expressed as the percentage of rosiglitazone depletion in the presence of the inhibitor compared with the control (with no inhibition). The mean of rosiglitazone metabolism in the presence of each inhibitor was compared with the control using the Student’s t test. The correlation analyses between the total activity of rosiglitazone metabolism and the activity of individual P450 enzymes (provided by the manufacturer) were conducted using Pearson correlation analysis. All statistical analyses were performed using Prism software (GraphPad Prism v5.01, GraphPad Software, Inc. CA, USA).
3 Results
3.1 Kinetics of Rosiglitazone Metabolism
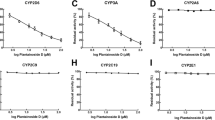
Rosiglitazone metabolism was NADPH dependent, and the enzyme kinetic model of Michaelis–Menten with substrate inhibition best described the kinetics of rosiglitazone metabolism in the majority of human liver microsome samples (Fig. 1). The kinetic parameters obtained from best model were as follows: K m between 17.1 and 29.8 (25 ± 5.57) µM, V max between 0.89 and 3.12 (1.64 ± 0.98) nmol·mg−1·min−1, and a K i between 4.25 and 234 (76 ± 91.9) µM. In only one sample (JOX) did the kinetics of rosiglitazone metabolism not converge in the MM,IS kinetic model, and it was best described with allosteric kinetics with a positive cooperativity (allosteric coefficient of n = 5.87), a K′ of 912 µM, and a V max of 0.27 nmol·mg−1·min−1. The enzyme kinetic parameters of the metabolism of rosiglitazone in individual human liver microsomes are presented in Table 2 (see also Fig. 1).
3.2 Impact of Inhibition on Rosiglitazone Metabolism Kinetics
The inhibition with montelukast or sulfaphenazole resulted in a slight decrease in the reaction velocity of rosiglitazone metabolism in all samples across the full substrate concentration range (between 0.5 and 500 µM) (Fig. 2). The kinetic parameters obtained for the best fitted enzyme kinetic model for the metabolism of rosiglitazone in individual human liver microsomes in the presence of the CYP2C8 or CYP2C9 inhibitors are provided in Table 2. In the presence of montelukast, a K m between 12.3 and 82.8 (33 ± 33.5) µM, a V max between 0.66 and 3.91 (1.72 ± 1.5) nmol·mg−1·min−1, and a K i between 9.1 and 859 (311 ± 388) µM were observed. In the presence of sulfaphenazole, the Km was in the range of 22.2–55.1 (39.1 ± 15) µM, V max was in the range of 1.08–5.4 (2.2 ± 1.8) nmol·mg−1·min−1, and K i was between 3.8 and 30.2 (15.8 ± 12.4) µM. For sample JOX, in the presence of montelukast, the contributing enzyme showed a positive cooperativity (allosteric coefficient of n = 4.36), with the K′ of 1667 µM and a V max of 0.24 nmol·mg−1·min−1. In the presence of sulfaphenazole, the kinetic parameters obtained from the allosteric kinetic model for sample JOX was allosteric coefficient of n = 4.31, a V max of 0.19 nmol·mg−1·min−1, and a K′ of 205 µM (see Fig. 2).
3.3 Impact of Inhibition on Rosiglitazone Metabolism at 10 µM
The results of inhibition experiments at a single 10 µM rosiglitazone concentration are presented in Fig. 3. Among the inhibitors tested, only quinidine (CYP2D6, 14% inhibition) and furafylline (CYP1A2, 18% inhibition) did not significantly (P > 0.05) decrease rosiglitazone metabolism compared with the rosiglitazone metabolism with no inhibition. In the presence of montelukast (CYP2C8, 49% inhibition), sulfaphenazole (CYP2C9, 30% inhibition at 2.5 µM and 42% inhibition at 10 µM), ketoconazole (CYP3A4, 52% inhibition), troleandomycin (CYP3A4, 41% inhibition), 13-cis retinoic acid (CYP2C8, 69% inhibition), and diethyldithiocarbamate (CYP2E1, 41% inhibition), and in the combination of both montelukast and sulfaphenazole (CYP2C8 and CYP2C9, 71% inhibition), there was a significant (P < 0.05) decrease in rosiglitazone metabolism compared with the no inhibition condition (see Fig. 3).
Inhibition of rosiglitazone metabolism (10 µM) in human liver microsomes in the presence of CYP inhibitors. *Significant difference (P < 0.05) in comparison with control. 13-cis 13-cis retinoic acid (CYP2C8) 142 µM, Die diethyldithiocarbamate (CYP2E1) 40 µM, Fur furafylline (CYP1A2) 10 µM, Ket ketoconazole (CYP3A4) 1 µM, Mon montelukast (CYP2C8) 0.02 µM, Mon + Sul montelukast (CYP2C8) 0.02 µM in combination with sulfaphenazole (CYP2C9) 2.5 µM, Qui quinidine (CYP2D6) 1 µM, Sul sulfaphenazole (CYP2C9) 2.5 µM or 10 µM, Tro troleandomycin (CYP3A4) 40 µM
4 Discussion
This study describes for the first time the evaluation of rosiglitazone metabolism in human liver microsomes using the substrate depletion method. Previous studies conducted using the product formation method reported that para-hydroxylation and N-demethylation are the main pathways of rosiglitazone metabolism in human liver microsomes, with the major contribution of CYP2C8 and a minor contribution of CYP2C9 [6]. In this study, we took a very different approach (substrate depletion) to Baldwin’s group (formation of metabolites) to assessing rosiglitazone metabolism and showed that further to the contribution of CYP2C8 andCYP2C9, CYP3A4 and CYP2E1 also contribute to the metabolism of rosiglitazone. The enzyme kinetics reported earlier using the product formation method for para-hydroxylation or N-demethylation of rosiglitazone in three human liver microsomes and two CYP2C8 Supersomes™ demonstrated ordinary Michaelis–Menten kinetics [6]. However, in the present study, using the substrate depletion method (which considers the sum of the activity of all the contributing enzymes), rosiglitazone metabolism exhibited substrate inhibition kinetics in four samples and allosteric kinetics in one sample. This indicates that the kinetics of rosiglitazone metabolism in total (the sum of the total metabolism via all the contributing enzymes) is different to the kinetics of the para-hydroxylation and N-demethylation pathways alone.
The mean V max obtained using the substrate depletion method in the present study was only 0.59-fold of the mean value reported by Baldwin et al. [6]. This was unexpected, since Baldwin and colleagues [6] only measured the formation of para-hydroxylation and N-demethylation metabolites, accounting for only approximately 60% of total oxidative metabolism [4], whereas the substrate depletion method used in the present study represents the sum of the total oxidative metabolism via all the contributing enzymes. One possibility for this apparent discrepancy is that the differences in the V max for rosiglitazone metabolism in our study and that of Baldwin and colleagues were due to inter-individual differences in the capacity to metabolize rosiglitazone. It has been demonstrated that the pharmacokinetics of rosiglitazone [clearance (Cl) and terminal half-life (t ½)] demonstrate inter-individual differences up to 2.6-fold [4]. Furthermore, the range of rosiglitazone para-hydroxylation was from 1 to 38 nmol·mg−1·h−1 and its N-demethylation was from 0.8 to 31 nmol·mg−1·h−1 at the single substrate concentration of 10 µM in liver microsomes obtained from 47 individuals [6], resulting in a combined activity in the order of 2–70 nmol·mg−1·h−1. The rosiglitazone metabolism in our study at the same concentration ranged from 11 to 47 nmol·mg−1·h−1 in the five samples we had access to, which is within the Baldwin’s results range [6]. This suggests that the lower V max in our study may have simply been due to the microsomes used in our experiments being derived from individuals with a lower capacity for rosiglitazone metabolism than the three samples used by Baldwin for the full kinetic experiments which determined V max.
The contribution of the CYP2C8 enzyme to the metabolism of rosiglitazone in human liver microsomes was demonstrated in both sets of experiments in our study. In the presence of montelukast over a substrate range of 0.5–500 µM, the K dep and therefore the initial velocity of rosiglitazone metabolism decreased in all individual samples and there were changes in the kinetic parameters of rosiglitazone metabolism. The decrease in the initial velocity of rosiglitazone metabolism at a substrate concentration of 10 µM (the same concentration as in Baldwin’s study [6]) was also observed in all of the samples. Furthermore, in the other set of inhibition studies at a rosiglitazone concentration of 10 µM, the percentage of rosiglitazone depletion in the presence of montelukast or 13-cis retinoic acid was significantly lower than without inhibition. These findings are in agreement with previous studies where the contribution of CYP2C8 in the metabolism of rosiglitazone, using the drugs believed to be mainly specific for CYP2C8, fluvoxamine [15] and trimethoprim [16], and the CYP2C8 inducer, rifampin [8, 16], has been demonstrated. However, our study demonstrated that CYP2C9, CYP3A4, and CYP2E1 also contribute to the metabolism of rosiglitazone.
In humans, CYP2C8 is genetically polymorphic and the CYP2C8*3 genotype confers altered metabolic activity of the CYP2C8 enzyme. The CYP2C8*3 genotype is associated with decreased oxidative capacity in vitro for paclitaxel [17]. In vivo, the CYP2C8*3 genotype has been associated with increased metabolism of repaglinide and pioglitazone and also decreased metabolism of R-ibuprofen [16, 18, 19]. For rosiglitazone, two clinical studies showed increased clearance of rosiglitazone in association with CYP2C8*3 genotype [18, 20]; however, two other studies have shown no significant association between CYP2C8*3 genotype and the pharmacokinetics of rosiglitazone [14, 21]. Therefore, this again suggests that CYP2C8 is not the only enzyme involved in the overall rosiglitazone metabolism.
The contribution of CYP2C9 to the metabolism of rosiglitazone was demonstrated in both sets of experiments. In the presence of sulfaphenazole over a substrate range of 0.5–500 µM, there were changes in the kinetic parameters of rosiglitazone metabolism. It has been reported that CYP2C9 has a minor contribution to the para-hydroxylation (~15%) and N-demethylation (~30%) of rosiglitazone in human liver microsomes [6]. However, in our inhibition studies at a substrate concentration of 10 µM, the contribution of CYP2C9 in the metabolism of rosiglitazone was significant at the tested inhibitor concentrations, 30% inhibition at 2.5 µM and 42% inhibition at 10 µM. These findings suggest that while CYP2C9 might confer a minor contribution to para-hydroxylation and N-demethylation of rosiglitazone, it contributes almost the same as CYP2C8 to rosiglitazone metabolism overall.
The substrate depletion method does not provide information on metabolic pathways of a substrate; however, it can clearly demonstrate the contribution of CYP enzymes in the overall metabolism of the substrate. In this study, by using the substrate depletion method, we further demonstrated that other CYP enzymes, including CYP3A4 and CYP2E1, play a significant role in rosiglitazone metabolism. In an in vivo study in humans, ketoconazole increased the mean area under the plasma concentration–time curve (AUC) of rosiglitazone by 47% and also increased the peak plasma concentration of rosiglitazone by 17% [8]; however, the authors concluded that the observed rosiglitazone inhibition was due to the inhibition of CYP2C8 or CYP2C9 by ketoconazole, despite the fact that ketoconazole is also a potent inhibitor of CYP3A4. It has been reported that ketoconazole (as a selective CYP3A4 inhibitor with the K i of 0.02–0.11 µM) can inhibit CYP2C8 activity with the K i of 2.5 µM [7]. However, in our inhibition studies, the ketoconazole concentration was 1 µM (<K i for CYP2C8 inhibition), and therefore, the observed inhibition appears to be due to selective CYP3A4, rather than CYP2C8, inhibition. Importantly, this is the same concentration used by Baldwin et al. [6] in their work examining the formation of the N-desmethyl and para-hydroxy metabolites. They observed a 15% decrease in N-demethylation and about 3% decrease in para-hydroxylation of rosiglitazone, while we observed a 52% decrease in total rosiglitazone metabolism. Furthermore, using another selective inhibitor of CYP3A4, troleandomycin (40 µM), we demonstrated a 40% inhibition of rosiglitazone metabolism. The inhibition of CYP2C8 in the presence of 50 µM troleandomycin, which is slightly higher (1.2-fold) than that used in our work, has been reported to be less than 10% [7]. Therefore, these combined results confirm the significant contribution of CYP3A4 in rosiglitazone metabolism, despite its relatively minor role in forming the N-desmethyl and para-hydroxy metabolites.
Our results also demonstrated a ~41% inhibition of rosiglitazone metabolism in the presence of a selective inhibitor of CYP2E1, diethyldithiocarbamate. Interestingly, Baldwin et al. in their work demonstrated a significant correlation between N-demethylation of rosiglitazone with lauric acid ω-1 hydroxylase (CYP2E1) activity in 47 human liver microsomes [6]. However, they did not further investigate the contribution of CYP2E1 in rosiglitazone metabolism. Therefore, this is the first study that has demonstrated the significant contribution of CYP2E1 in the metabolism of rosiglitazone in human liver microsomes.
Since rosiglitazone has negligible renal clearance, it might be more susceptible to metabolic drug interactions. Databases for prescribing rosiglitazone, such as the Medical Information Management System (MIMS), indicate that CYP2C8 is the only responsible enzyme for rosiglitazone metabolism. In contrast, this study provides a more comprehensive picture of rosiglitazone metabolism and suggests that, in addition to CYP2C8, the contribution of CYP2C9, CYP3A4, and CYP2E1 to the metabolism of rosiglitazone is significant. The work presented here highlights the potential for drug–drug interactions to be underestimated when the assessment is based only on product formation metabolism studies, since these studies may not provide information on important metabolic pathways/enzymes that contribute to overall drug metabolism. Understanding the complete metabolism information of rosiglitazone will assist in improving the current therapeutic strategies and also in better managing its use in regard to polypharmacy and pharmacogenetics.
References
Camp H. Thiazolidinediones in diabetes: current status and future outlook. Curr Opin Investig Drugs. 2003;4:406.
Iwamoto Y, Kuzuya T, Matsuda A, Awata T, Kumakura S, Inooka G, Shiraishi I. Effect of new oral antidiabetic agent cs-045 on glucose tolerance and insulin secretion in patients with NIDDM. Diabetes Care. 1991;14:1083–6.
Suter S, Nolan J, Wallace P, Gumbiner B, Olefsky J. Metabolic effects of new oral hypoglycemic agent cs-045 in NIDDM subjects. Diabetes Care. 1992;15:193–203.
Cox PJ, Ryan DA, Hollis FJ, Harris AM, Miller AK, Vousden M, Cowley H. Absorption, disposition, and metabolism of rosiglitazone, a potent thiazolidinedione insulin sensitizer, in humans. Drug Metab Dispos. 2000;28:772–80.
Bolton GC, Keogh JP, East PD, Hollis FJ, Shore AD. The fate of a thiazolidinedione antidiabetic agent in rat and dog. Xenobiotica. 1996;26:627–36.
Baldwin SJ, Clarke SE, Chenery RJ. Characterization of the cytochrome p450 enzymes involved in the in vitro metabolism of rosiglitazone. Br J Clin Pharmacol. 1999;48:424–32.
Ong CE, Coulter S, Birkett DJ, Bhasker CR, Miners JO. The xenobiotic inhibitor profile of cytochrome P4502C8. Br J Clin Pharmacol. 2000;50:573–80.
Park JY, Kim KA, Shin JG, Lee KY. Effect of ketoconazole on the pharmacokinetics of rosiglitazone in healthy subjects. Br J Clin Pharmacol. 2004;58:397–402.
Siller M, Anzenbacher P, Anzenbacherova E, Dolezal K, Popa I, Strnad M. Interactions of olomoucine II with human liver microsomal cytochromes p450. Drug Metab Dispos. 2009;37:1198–202.
Khojasteh SC, Prabhu S, Kenny JR, Halladay JS, Lu AYH. Chemical inhibitors of cytochrome p450 isoforms in human liver microsomes: a re-evaluation of p450 isoform selectivity. Eur J Drug Metab Pharmacokinet. 2011;36:1–16.
Walsky RL, Obach RS. Validated assays for human cytochrome p450 activities. Drug Metab Dispos. 2004;32:640–7.
Newton DJ, Wang RW, Lu A. Cytochrome p450 inhibitors. Evaluation of specificities in the in vitro metabolism of therapeutic agents by human liver microsomes. Drug Metab Dispos. 1995;23:154–8.
Chauret N, Gauthier A, Nicoll-Griffith DA. Effect of common organic solvents on in vitro cytochrome P450-mediated metabolic activities in human liver microsomes. Drug Metab Dispos. 1998;26:1–4.
Bazargan M, Muhlhausler BS, Morrison JL, McMillen IC, Davey AK, Foster DJR. Simple HPLC method for determination of rosiglitazone in sheep plasma and amniotic fluid and its application in a pregnant sheep model. J Pharm Biomed Anal. 2011;55:360–5.
Pedersen RS, Damkier P, Brosen K. The effects of human CYP2C8 genotype and fluvoxamine on the pharmacokinetics of rosiglitazone in healthy subjects. Br J Clin Pharmacol. 2006;62:682–9.
Niemi M, Backman JT, Neuvonen PJ. Effects of trimethoprim and rifampin on the pharmacokinetics of the cytochrome P450 2C8 substrate rosiglitazone. Clin Pharmacol Ther. 2004;76:239–49.
Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter JG, Burhan I, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenet Genom. 2001;11:597–607.
Aquilante CL, Kosmiski LA, Bourne DW, Bushman LR, Daily EB, Hammond KP, Hopley CW, Kadam RS, Kanack AT, Kompella UB. Impact of the CYP2C8* 3 polymorphism on the drug–drug interaction between gemfibrozil and pioglitazone. Br J Clin Pharmacol. 2013;75:217–26.
Martínez C, García-Martín E, Blanco G, Gamito FJ, Ladero JM, Agúndez JA. The effect of the cytochrome P450 CYP2C8 polymorphism on the disposition of (R)-ibuprofen enantiomer in healthy subjects. Br J Clin Pharmacol. 2005;59:62–9.
Kirchheiner J, Thomas S, Bauer S, Tomalik-Scharte D, Hering U, Doroshyenko O, Jetter A, Stehle S, Tsahuridu M, Meineke I. Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Br J Clin Pharmacol. 2006;80:657–67.
Hruska M, Cheonge JA, Amico J, Langaee T, Ferrell R, Fitzgerald S, Frye R. The effect of CYP2C8 genotype on rosiglitazone pharmacokinetics. Br J Clin Pharmacol. 2005;. doi:10.1016/j.clpt.2004.12.029.
Author Contributions
Participation in research design: Maryam Bazargan and David Foster. Conducted experiments: Maryam Bazargan. Performed data analysis: Maryam Bazargan and David Foster. Wrote the manuscript: Maryam Bazargan. Contributed to the writing of the manuscript and interpretation of the results: Maryam Bazargan, David Foster, Andrew Davey, and Beverly Muhlhausler.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was funded by University of South Australia, Ph.D. research funding. Maryam Bazargan, David Foster, Beverly Muhlhausler, and Andrew Davey state that they have no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bazargan, M., Foster, D.J.R., Davey, A.K. et al. Rosiglitazone Metabolism in Human Liver Microsomes Using a Substrate Depletion Method. Drugs R D 17, 189–198 (2017). https://doi.org/10.1007/s40268-016-0166-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40268-016-0166-4